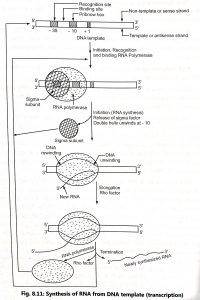
Transcription involves three different stages in prokaryotes such as initiation, elongation and termination. RNA is synthesized by a single RNA polymerase enzyme which contains multiple polypeptide subunits. RNA polymerase of Escherichia coli is a complex holoenzyme with five polypeptide subunits (2α, 1β, 1β’, 1σ factor). The sigma (a) factor may dissociate from the other subunits to form core enzyme (2α, 1β, iβ’).
Initiation: The enzyme RNA polymerase plays a major role in recognition and binding of initiation site. The specific region on the DNA where the enzyme binds is known as promoter region. Promoter region contains specific DNA sequences that acts as points of attachment for the RNA polymerase. In Escherichia coli, two sequence elements recognized by the RNA polymerase known as the – 10 sequence and -35 sequence. The Pribnow’s box (TATA box), whereas 35 region is called the recognition site. The Pribnow’s box is found as a part of all prokaryotic promoters. It consists of 6 nucleotide basis (TATAAT), located on the left side about 10 bases away from the starting point of transcription (Fig. 8.11). The double helix DNA partially dissociates at Pribnow’s box (- 10 box), which is rich in weak A-T bonds to give an open promoter complex. This open complex then allows tight binding of the RNA polymerase with subsequent initiation of RNA synthesis.
Elongation: RNA is synthesized from 5′ end to 3′ end and (5′ -→ 3′) antiparallel to the DNA template. Chain elongation occurs by core enzyme that move along the DNA template.The region of template that has been transcribed regains its double helical form behind the bubble and the next region of DNA which is ta be transcribed unwinds. The unwound area contains the newly synthesized RNA base-paired with the template DNA strand and extends over 12 to 17 bases. The newly formed RNA transcript consists of a triphosphate group at 5 and free – OH group at 3′ end.
Termination: The process of transcription stops by termination signals. There are two types of mechanisms that brings about termination such as rho-independent termination and rho-dependent termination. The rho-independent termination is brought about by the formation of hairpins of newly synthesized RNA. This occurs due to the presence of palindromes. It synthesises an RNA transcript that becomes folded to form a stem and loop structure of about 20 bases. This step-loop structure causes RNA polymerase to pause a disrupt the RNA-DNA hybrid at 5 end.
Rho-dependent termination requires the Rho protein (factor) encode by rho-gene. It binds to the growing RNA of weakly to DNA, and in the bound state it acts as ATPase and terminates transcription. The termination of transcription involves the release of the transcript and the core enzyme which may then reassociate with the sigma subunit and it starts another round of transcription.