Contents:
- 1. Binary Fission: The Primary Mode of Bacterial Reproduction
- 2. Genetic Exchange Mechanisms in Bacteria
- 3. Endospore Formation
- 4. Budding
- 5. Multiple Fission
- 6. Reproduction in Filamentous Bacteria
- 7. Cell Division in Unusual Bacteria
- 8. Molecular Mechanisms of Cell Division
- 9. Environmental Factors Affecting Bacterial Reproduction
- 10. Bacterial Growth Curve
- 11. Quorum Sensing and Population-Level Reproduction Control
- Frequently Asked Questions (FAQ)
- References
Bacteria represent one of the most ancient and successful forms of life on Earth, with remarkable reproductive capabilities that have enabled them to colonize virtually every habitat on the planet. Their reproductive mechanisms are both elegant and efficient, allowing for rapid population growth under favorable conditions. This article explores the various mechanisms through which bacteria reproduce and propagate their genetic material.
1. Binary Fission: The Primary Mode of Bacterial Reproduction
Binary fission is the predominant method of reproduction in bacteria, representing an asexual process that creates genetically identical daughter cells.
The Process of Binary Fission
Binary fission occurs through several distinct phases:
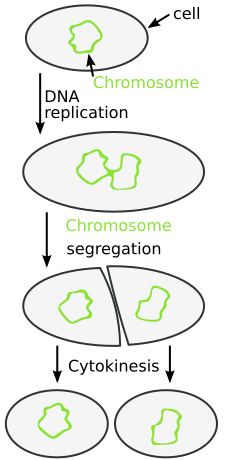
- DNA Replication: The circular bacterial chromosome replicates, beginning at the origin of replication (oriC) and proceeding bidirectionally until two complete chromosomes are formed.
- Cell Elongation: The cell grows to approximately twice its original size to accommodate the duplicated genetic material and cellular components.
- Chromosome Segregation: The replicated chromosomes move toward opposite poles of the cell, ensuring each daughter cell will receive a complete genome.
- Septum Formation: A cell wall grows inward from the cell membrane at the midpoint between the segregated chromosomes, forming a septum.
- Cell Division: The septum completes, physically separating the cytoplasm into two compartments and forming two distinct daughter cells.
Timing and Efficiency of Binary Fission
The rate of binary fission varies dramatically among bacterial species and is heavily influenced by environmental conditions. Under optimal laboratory conditions:
| Bacterial Species | Generation Time | Potential Population After 24 Hours (Starting with 1 Cell) |
|---|---|---|
| Escherichia coli | 20 minutes | Approximately 4.7 × 10^21 cells |
| Bacillus subtilis | 30 minutes | Approximately 2.8 × 10^14 cells |
| Mycobacterium tuberculosis | 24 hours | Approximately 16 cells |
| Treponema pallidum | 33 hours | Approximately 2 cells |
Control of Binary Fission
Several mechanisms regulate bacterial cell division:
- Min System: Proteins MinC, MinD, and MinE oscillate from pole to pole, inhibiting septum formation except at the midpoint of the cell.
- Nucleoid Occlusion: The protein Noc (in B. subtilis) or SlmA (in E. coli) prevents Z-ring formation over unsegregated chromosomes.
- FtsZ and the Z-Ring: FtsZ proteins polymerize to form a ring-like structure at the future division site, recruiting other division proteins.
- Environmental Sensing: Nutrient availability, temperature, and other environmental factors influence division rate through various signaling pathways.
2. Genetic Exchange Mechanisms in Bacteria
Although bacteria reproduce asexually, they possess several mechanisms for genetic exchange that contribute to genetic diversity and evolution.
Transformation
Transformation involves the uptake of naked DNA from the environment.
- Natural Competence: Some bacteria (like Streptococcus pneumoniae and Bacillus subtilis) naturally develop a physiological state called competence that allows them to take up DNA.
- The Process:
- Release of DNA from lysed cells into the environment
- Binding of DNA to receptors on the competent cell’s surface
- DNA fragmentation and transport of one strand into the cytoplasm
- Integration of the foreign DNA into the bacterial chromosome through homologous recombination
Conjugation
Conjugation is often described as bacterial “mating” and involves direct cell-to-cell contact.
- Key Components:
- F plasmid (fertility factor): Contains genes necessary for the conjugation process
- Pilus: A tubular structure that extends from donor to recipient, facilitating cell contact
- The Process:
- A donor cell (F+ or Hfr) makes contact with a recipient cell (F-) via a sex pilus
- The cells are drawn together, and a conjugation bridge forms
- DNA transfer occurs in a 5′ to 3′ direction through the bridge
- The recipient cell becomes F+ if the entire F plasmid is transferred
| Type of Donor Cell | Transferred Genetic Material | Outcome in Recipient |
|---|---|---|
| F+ | F plasmid | Recipient becomes F+ |
| Hfr (High frequency recombination) | Chromosomal DNA (with potential for F plasmid) | Recipient potentially receives new genes without becoming F+ |
| F’ | F plasmid carrying some bacterial genes | Recipient becomes F+ and receives additional bacterial genes |
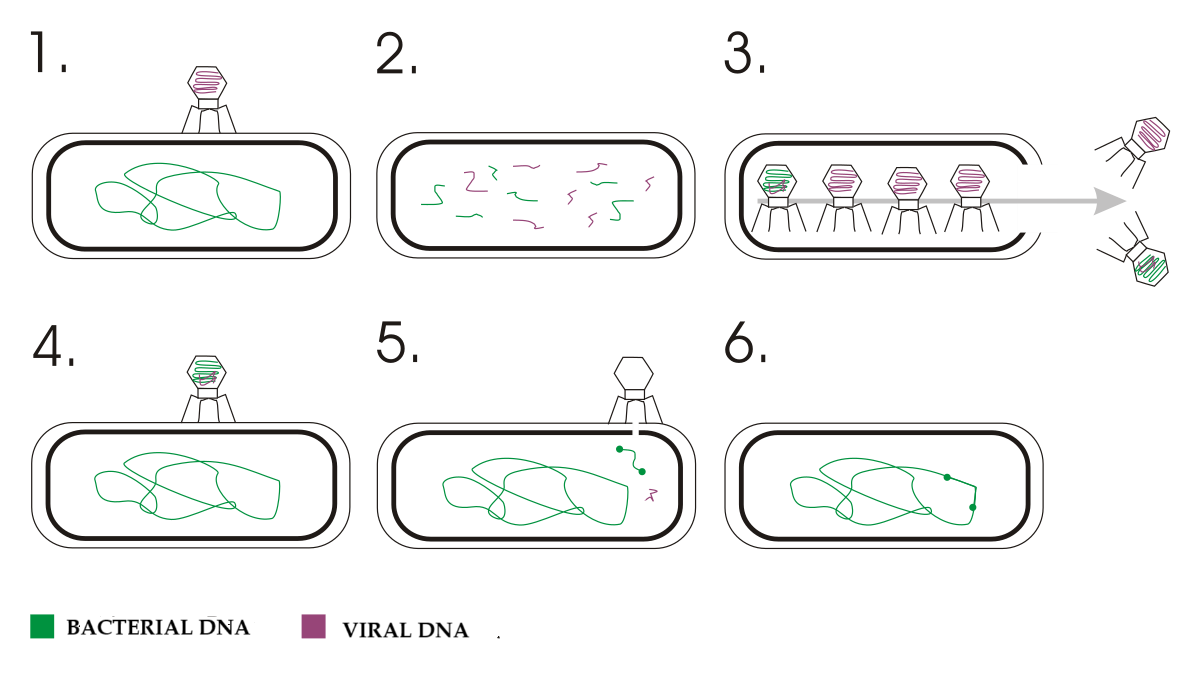
Transduction
Transduction involves the transfer of bacterial DNA via bacteriophages (viruses that infect bacteria).
- Types of Transduction:
- Generalized Transduction: Random bacterial DNA fragments are accidentally packaged into phage capsids during lytic cycle
- Specialized Transduction: Specific bacterial genes adjacent to the prophage integration site are packaged with phage DNA during imprecise excision of the prophage
- The Process of Generalized Transduction:
- Bacteriophage infects a donor bacterium
- During viral replication, host DNA is degraded and phage DNA is replicated
- Occasionally, bacterial DNA fragments are mistakenly packaged into phage capsids
- The phage lyses the donor cell and infects a recipient bacterium
- Bacterial DNA is injected into the recipient instead of phage DNA
- The bacterial DNA can recombine with the recipient’s chromosome
3. Endospore Formation
Some Gram-positive bacteria (notably Bacillus and Clostridium species) can form highly resistant structures called endospores in response to adverse environmental conditions.
The Process of Sporulation
Sporulation proceeds through several distinct stages:
- Stage I: DNA replication and axial filament formation
- Stage II: Asymmetric cell division creating a forespore and mother cell
- Stage III: Engulfment of the forespore by the mother cell
- Stage IV: Cortex formation between membranes
- Stage V: Coat protein deposition
- Stage VI: Maturation of the spore and development of resistance properties
- Stage VII: Lysis of the mother cell and release of the mature spore
Endospore Resistance Properties
| Resistance Factor | Protective Mechanism | Degree of Resistance |
|---|---|---|
| Heat | Dehydrated core, calcium-dipicolinic acid complex | Can survive boiling for hours |
| Radiation | DNA bound to small acid-soluble proteins (SASPs) | Up to 10 times more resistant than vegetative cells |
| Chemicals | Impermeable outer coat layers | Resistant to many disinfectants |
| Desiccation | Low water content in core | Can remain viable for thousands of years |
| Enzymes | Multiple protective layers | Resistant to enzymatic degradation |
Germination
When favorable conditions return, endospores can germinate to produce a vegetative cell:
- Activation: Often requires specific triggers such as heat shock
- Germination: Involves rehydration, degradation of cortex, and resumption of metabolism
- Outgrowth: The new vegetative cell emerges from the spore coat and resumes growth
4. Budding
Some bacteria reproduce by budding, a process where a small outgrowth (bud) forms on the parent cell, grows, and eventually separates.
- Examples: Hyphomonas, Caulobacter crescentus, and some cyanobacteria
- The Process:
- DNA replication in the parent cell
- Formation of a bud at a specific location on the cell surface
- One copy of the chromosome moves into the developing bud
- The bud grows until it reaches a certain size
- Separation of the bud from the parent cell
5. Multiple Fission
Multiple fission involves a single bacterial cell dividing into multiple offspring simultaneously.
- Examples: Some cyanobacteria, particularly those forming filaments
- The Process:
- Multiple rounds of DNA replication without cell division
- Growth of the parent cell
- Formation of multiple septa
- Simultaneous division into multiple daughter cells
6. Reproduction in Filamentous Bacteria
Filamentous bacteria form long chains or filaments of cells and have specialized reproductive strategies.
Fragmentation
- Occurs when filaments break into smaller pieces, each capable of growing into a new filament
- Common in filamentous cyanobacteria and actinomycetes
Specialized Reproductive Structures
- Conidia: Asexual spores formed by some actinomycetes (e.g., Streptomyces)
- Hormogonia: Short motile filaments that detach from the parent filament in some cyanobacteria
- Akinetes: Specialized resistant cells formed by some cyanobacteria in response to unfavorable conditions
7. Cell Division in Unusual Bacteria
Not all bacteria conform to the typical binary fission model, with some exhibiting unique reproductive strategies.
Baeocytes in Cyanobacteria
- Large cells divide internally to produce multiple small cells (baeocytes)
- Common in Pleurocapsalean cyanobacteria
FtsZ-Independent Division
- Some bacteria, like members of the Planctomycetes-Verrucomicrobia-Chlamydiae superphylum, divide without using the FtsZ protein
- These bacteria often use alternative cytoskeletal proteins for cell division
Longitudinal Fission
- Some spiral-shaped bacteria, like certain spirochetes, divide longitudinally rather than transversely
- The division plane runs along the long axis of the cell
8. Molecular Mechanisms of Cell Division
The molecular machinery orchestrating bacterial cell division involves dozens of proteins working in coordinated fashion.
The Divisome Complex
- Central Component: FtsZ (prokaryotic tubulin homolog) forms the Z-ring
- Early Division Proteins: FtsA, ZipA, ZapA, ZapB, ZapC, ZapD stabilize the Z-ring
- Late Division Proteins: FtsK, FtsQ, FtsL, FtsB, FtsW, FtsI, FtsN contribute to septum formation and cell wall synthesis
Chromosome Segregation
- ParABS System: Ensures proper distribution of replicated chromosomes
- ParB proteins bind to parS sites near the origin of replication
- ParA proteins help move the ParB-bound DNA toward cell poles
- SMC Complex: Structural maintenance of chromosomes proteins help organize and compact the chromosome
Cell Wall Synthesis During Division
- Peptidoglycan Synthesis: FtsI (PBP3) and other penicillin-binding proteins synthesize new peptidoglycan at the division site
- Autolysins: Enzymes that cleave existing peptidoglycan bonds to allow for cell wall remodeling during division
9. Environmental Factors Affecting Bacterial Reproduction
Bacterial reproduction rates are profoundly influenced by environmental conditions.
Temperature Effects
| Temperature Range | Effect on Reproduction |
|---|---|
| Below minimum growth temperature | Metabolic reactions too slow for reproduction |
| Near optimal temperature | Maximum reproduction rate |
| Above optimal temperature | Protein denaturation, decreased reproduction |
| Lethal high temperature | No reproduction, cell death |
Nutrient Availability
- Carbon Source: Primary energy source for most bacteria, essential for cell building blocks
- Nitrogen Source: Required for protein and nucleic acid synthesis
- Phosphorus: Essential for DNA, RNA, and energy metabolism (ATP)
- Micronutrients: Trace elements needed as enzyme cofactors
pH and Osmotic Pressure
- Most bacteria reproduce optimally within specific pH ranges
- Extremes in osmotic pressure can cause plasmolysis or cell lysis, preventing reproduction
Oxygen Requirements
| Oxygen Requirement | Bacterial Type | Example Organisms |
|---|---|---|
| Obligate aerobes | Require oxygen | Pseudomonas aeruginosa |
| Microaerophiles | Require low oxygen concentrations | Campylobacter jejuni |
| Facultative anaerobes | Can grow with or without oxygen | Escherichia coli |
| Aerotolerant anaerobes | Don’t use oxygen but tolerate its presence | Streptococcus pyogenes |
| Obligate anaerobes | Cannot tolerate oxygen | Clostridium botulinum |
10. Bacterial Growth Curve
The bacterial growth curve describes population changes over time in a closed culture system.
Phases of the Growth Curve
- Lag Phase:
- Cells adapt to new environment
- Minimal reproduction
- Active metabolism and protein synthesis
- Duration varies based on previous growth conditions and new environment
- Exponential (Log) Phase:
- Cells divide at constant rate
- Population doubles at regular intervals
- Maximum metabolic activity
- Most susceptible to antibiotics targeting active processes
- Stationary Phase:
- Population growth equals death rate
- Nutrient depletion and waste accumulation
- Expression of stress response genes
- Morphological and physiological changes for survival
- Death (Decline) Phase:
- Death rate exceeds reproduction rate
- Accumulation of toxic waste products
- Nutrient exhaustion
- Some cells may enter viable but non-culturable state
Mathematical Representation of Bacterial Growth
During exponential phase, bacterial growth follows the equation: N = N₀ × 2^n
Where:
- N = final number of cells
- N₀ = initial number of cells
- n = number of generations
11. Quorum Sensing and Population-Level Reproduction Control
Bacteria can coordinate population-wide behaviors through chemical communication systems known as quorum sensing.
Mechanism of Quorum Sensing
- Autoinducers: Small signaling molecules produced and detected by bacteria
- Concentration Dependence: Signal concentration increases with population density
- Threshold Response: When autoinducer concentration reaches a threshold, gene expression changes
Quorum Sensing Systems
| System Type | Autoinducer | Typical in | Functions Regulated |
|---|---|---|---|
| AHL system | Acyl-homoserine lactones | Gram-negative bacteria | Biofilm formation, virulence, bioluminescence |
| Peptide system | Oligopeptides | Gram-positive bacteria | Competence, sporulation, virulence |
| AI-2 system | Furanosyl borate diester | Both Gram-positive and Gram-negative | Interspecies communication |
Population-Level Effects on Reproduction
- Biofilm Formation: Quorum sensing regulates the transition from planktonic to biofilm growth
- Sporulation: In Bacillus subtilis, population density signals influence sporulation decisions
- Growth Rate Modulation: Some species adjust growth rates based on population density
Frequently Asked Questions (FAQ)
Q1: Why do bacteria primarily reproduce asexually rather than sexually?
A: Asexual reproduction through binary fission allows bacteria to reproduce rapidly and efficiently when conditions are favorable. A single bacterial cell can quickly give rise to billions of genetically identical cells, which is advantageous for colonizing new environments. While bacteria lack true sexual reproduction, they have evolved alternative mechanisms for genetic exchange (transformation, conjugation, and transduction) that provide many of the evolutionary benefits of sexual reproduction without requiring the complex cellular machinery or time investment of meiosis and fertilization.
Q2: Can bacteria reproduce indefinitely under the right conditions?
A: Theoretically, bacteria can reproduce indefinitely under ideal conditions, as they don’t undergo programmed senescence like many eukaryotic cells. However, in practice, even in carefully controlled laboratory conditions, bacteria eventually accumulate mutations and other forms of damage that can affect reproduction. Additionally, bacteria in natural environments face resource limitations, competition, predation, and environmental fluctuations that prevent unlimited growth.
Q3: How do antibiotics target bacterial reproduction?
A: Different classes of antibiotics target various aspects of bacterial reproduction:
- Cell Wall Synthesis Inhibitors (penicillins, cephalosporins): Interfere with peptidoglycan synthesis, causing cell lysis during division
- Protein Synthesis Inhibitors (tetracyclines, aminoglycosides): Prevent production of essential proteins needed for reproduction
- DNA Replication Inhibitors (fluoroquinolones): Block DNA replication by inhibiting DNA gyrase and topoisomerase IV
- RNA Synthesis Inhibitors (rifampin): Inhibit bacterial RNA polymerase, preventing transcription of genes needed for reproduction
Q4: Do all bacteria reproduce at the same rate?
A: No, reproduction rates vary dramatically among bacterial species. Factors influencing reproduction rate include:
- Intrinsic Factors: Genome size, metabolic efficiency, cell structure
- Environmental Factors: Temperature, nutrient availability, oxygen levels, pH
- Growth Strategy: Some bacteria are adapted for rapid growth in resource-rich environments (r-strategists), while others are adapted for slower growth and persistence in stable environments (K-strategists)
Q5: How is bacterial reproduction different from eukaryotic cell division?
A: Key differences include:
- Bacterial DNA is typically circular and exists in the cytoplasm without a nuclear membrane
- Bacteria lack mitotic spindles and centrosomes
- Bacterial chromosome segregation uses different mechanisms than eukaryotic mitosis
- Bacterial cell division uses FtsZ (a tubulin homolog) rather than actin-myosin contractile rings
- Bacteria reproduce much more rapidly than most eukaryotic cells
- Most bacteria have a single chromosome rather than multiple chromosomes
Q6: Can bacteria reproduce through mechanisms other than those described in this article?
A: The mechanisms described in this article cover the majority of known bacterial reproductive strategies. However, the microbial world is incredibly diverse, and scientists continue to discover unusual reproductive mechanisms in newly characterized bacterial species. Some bacteria exhibit complex life cycles with multiple reproductive modes depending on environmental conditions. Additionally, within the broader category of prokaryotes, archaea may have unique reproductive strategies that differ from those used by bacteria.
References
- Angert, E. R. (2005). Alternatives to binary fission in bacteria. Nature Reviews Microbiology, 3(3), 214-224. https://www.nature.com/articles/nrmicro1096
- Adams, D. W., & Errington, J. (2009). Bacterial cell division: assembly, maintenance and disassembly of the Z ring. Nature Reviews Microbiology, 7(9), 642-653. https://www.nature.com/articles/nrmicro2198
- Higgins, D., & Dworkin, J. (2012). Recent progress in Bacillus subtilis sporulation. FEMS Microbiology Reviews, 36(1), 131-148. https://academic.oup.com/femsre/article/36/1/131/591308
- Thomas, C. M., & Nielsen, K. M. (2005). Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nature Reviews Microbiology, 3(9), 711-721. https://www.nature.com/articles/nrmicro1234
- Monod, J. (1949). The growth of bacterial cultures. Annual Review of Microbiology, 3(1), 371-394. https://www.annualreviews.org/doi/abs/10.1146/annurev.mi.03.100149.002103
- Bassler, B. L., & Losick, R. (2006). Bacterially speaking. Cell, 125(2), 237-246. https://www.cell.com/cell/fulltext/S0092-8674(06)00390-X
- Margolin, W. (2005). FtsZ and the division of prokaryotic cells and organelles. Nature Reviews Molecular Cell Biology, 6(11), 862-871. https://www.nature.com/articles/nrm1745
- Rolbicki, J. M., & Kepes, F. (2018). The bacterial nucleoid: A highly organized and dynamic structure. Journal of Cellular Biochemistry, 119(2), 1454-1465. https://onlinelibrary.wiley.com/doi/abs/10.1002/jcb.26363
- Kearns, D. B., & Losick, R. (2005). Cell population heterogeneity during growth of Bacillus subtilis. Genes & Development, 19(24), 3083-3094. http://genesdev.cshlp.org/content/19/24/3083.short
- Waters, C. M., & Bassler, B. L. (2005). Quorum sensing: cell-to-cell communication in bacteria. Annual Review of Cell and Developmental Biology, 21, 319-346. https://www.annualreviews.org/doi/abs/10.1146/annurev.cellbio.21.012704.131001
