INTRODUCTION
PARVOVIRUSES | AT A GLANCE
- The smallest known viruses.
- Virions in the range 18–26 nm in diameter.
- The name was derived from the Latin word parvus which means small.
- Family – Parvoviridae
- Subfamily – Parvovirinae (vertebrate viruses) and the Densovirinae (invertebrate viruses )
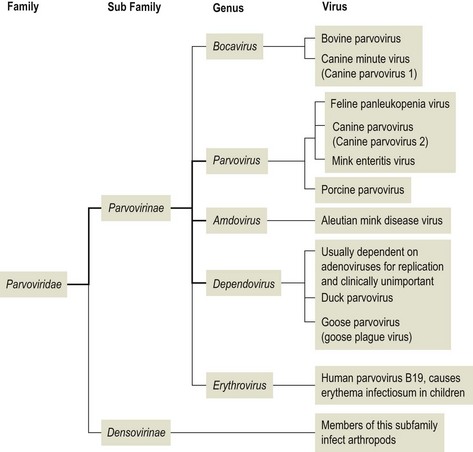
PARVOVIRUS VIRON STRUCTURE
- Simple structure with the ssDNA
- Genome enclosed within a capsid
- Icosahedral symmetry
- The capsid is built from 60 protein molecules
- The proteins are numbered in order of their size.
- Each of the protein species contains an eight-stranded β-barrel structure.
- The virion is roughly spherical.
- With surface protrusions and canyons
- A protrusion with a pore at the center is present at each of the vertices of the icosahedron.
GENOME
- Genomes composed of linear ssDNA
- Size range 4–6 kb
- Several short complementary sequences are present at each end of a DNA molecule.
- They can base pair to form a secondary structure.
- Inverted terminal repeats (ITRs) are present in some parvovirus genomes.
- The sequence at one end is complementary and in the opposite orientation to, the sequence at the other end.
- The ends have identical secondary structures as the sequences are complementary.
- Parvoviruses with ITRs during replication generate equal numbers of (+) and (−) strands of DNA.
- In the case of (-) DNA, the genes for non-structural proteins are towards the 3’end and the structural protein genes are towards the 5’end.

PARVOVIRUS REPLICATION
- Its small genome can encode only a few proteins.
- The virus depends on its host cell or another virus to provide important proteins
- Some of these proteins are available only during the S phase of the cell cycle during DNA synthesis.
- Hence, this restricts the opportunity for parvovirus replication to the S phase.
- The virion attaches to receptors on the surface of a host cell.
- Enters the cell by endocytosis and is released from the endosome into the cytoplasm.
- Associates with microtubules and is transported to a nuclear pore.
- The parvovirus virion is too small to pass through a nuclear pore.
- The capsid proteins of some parvoviruses undergo nuclear localization signals.
- The single-stranded virus genome is converted to dsDNA inside the nucleus by a cell DNA polymerase.
- As a result of base pairing, the ends of the genome are double-stranded.
- At the 3’end the –OH group acts as a primer to which the enzyme binds.
- The virus genes are transcribed by the cell RNA polymerase II.
- The primary transcript(s) undergo various splicing events.
- Two size classes of mRNA are produced.
- The non-structural proteins are encoded by larger mRNAs.
- The smaller mRNAs encode the structural proteins.
- The non-structural proteins are phosphorylated.
- The DNA is replicated by a mechanism called rolling-hairpin replication after the conversion of the ssDNA genome to dsDNA.
- This mechanism sets parvoviruses apart from other DNA viruses.
- Capsids are constructed from the structural proteins and filled by a copy of the virus genome, either a (+) DNA or a (−) DNA as required.
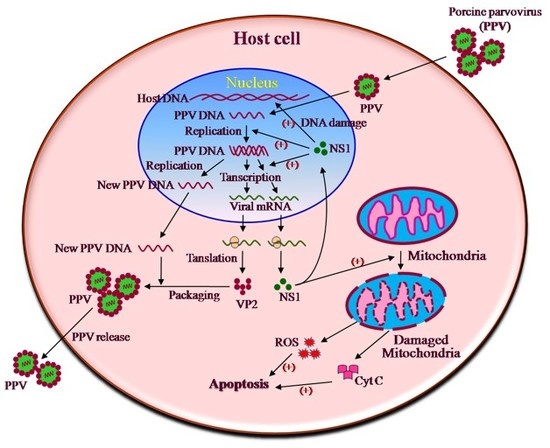
REFERENCE
- https://pubmed.ncbi.nlm.nih.gov/29261104/
- https://pubmed.ncbi.nlm.nih.gov/28410597/
- https://www.academia.edu/30888358/John_Carter_Virology_Principles_and_Applications.pdf
