Polymerase Chain Reaction (PCR)
PCR is an in-vitro method for producing large amounts of specific DNA fragments of defined length and sequence from a small amount of complex template, In this technique microgram quantities of DNA from picogram produce amounts of starting material, Target DNA, primers, polymerase and nucleotides are combined in a test tube for multiplication of genetic material.
DNA is amplified by polymerase chain reaction (Fig. 12.10) in an enzymatic reaction which undergoes multiple incubations at three different temperatures. Each PCR contains four important components.
DNA Template : Any source that contains one or more target DNA molecules to be amplified can be taken as template. RNA can also be used for PCR by first making a DNA copy using the enzyme reverse transcriptase.
Primers: Each PCR requires a pair of oligonucleotide primers, These are short single-stranded DNA molecules obtained by chemical synthesis. These primers are designed to anneal on opposite strands of target sequence so that they will be extended towards each other by addition of nucleotides.
DNA polymerase: The most commonly used enzyme in PCR is Tag DNA polymerase isolated from a thermostable bacterium called Thermus aquaticus. It survives at 95°C for 1 to 2 minutes and has a half life for more than 2 hours at same temperature. The DNA polymerase binds to a single-stranded DNA and synthesizes a new strand complementary to the original strand. The role of this enzyme in PCR is to copy DNA molecules.
Deoxynucleotide triphosphates: PCR requires four deoxynucleotide triphosphates, DNTPS (DATP, dGTP, DTTP, dCTP) which are used by the DNA polymerase as building blocks to synthesize new DNA.
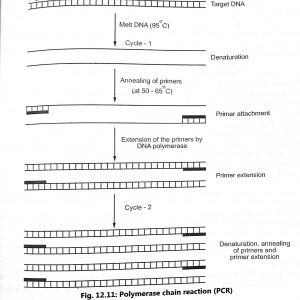
Polymerase chain reaction (PCR) involves three stages which are as follows (Fig. 12.11):
- Melting of DNA (95°C) to convert double stranded DNA to a single stranded DNA (denaturation).
- Promoting the primers (at 50 65°C) to attach themselves to either end of the target strip (annealing of primers).
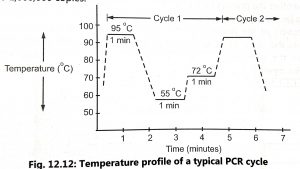
- Extension of the primers DNA polymerase to form new double-stranded DNA across the segment by sequential addition of deoxynucleotides (primer extension). When the temperature is again raised the new strands separate and the process begins again. Temperature profile of typical PCR cycle is shown in Fig. 12.12.
The oligonucleotide primers are designed to hybridise the region of DNA flanking a desired target gene sequence. The primers are then extended across the target sequence using DNA polymerase derived from Thermus aquaticus (Taq) in the presence of free deoxynucleotide triphosphate. These three steps constitute one cycle of the reaction. These steps are repeated by manipulating the temperature, by using the PCR machine, A cycle takes about 3 to 5 minutes and after 30 cycles (about 3 hours), a single copy of DNA can be multiplied into 1,000,000 copies.
RNA polymerase chain reaction is a modification of PCR technique which allows amplification beginning from an RNA template. Recent research in the field of polymerase chain reaction has led to the development of the techniques contributing to the effectiveness of the method. PCR is a highly versatile technique. Important variations of PCR are as follows:
Inverse PCR: PCR can be used to the amplification of those DNA sequences which are away from the primer and not of those which are flanked by the primer. The sequences to be amplified may be cloned in a vector and border sequence of the vector may be used as a primer in such a way that the polymerization proceeds in reverse direction.
Anchored PCR: In this method only one primer is used. One strand is copied first and then poly G tail is attached at the end of the newly synthesized strand. This allows the use of complementary homopolymer, poly-C, to be used as primer for copying the DNA single strands generated by PCR. It gives rise to the complete DNA duplex that can be amplified normally.
RT-PCR: Reverse transcription-mediated PCR includes a single application combining the process of CDNA synthesis (by reverse transcription) and PCR amplification. It can also be applied to double stranded CDNA also which is synthesized from mRNA using the enzyme reverse transcriptase. Thermostable enzyme rTth uses RNA templates from cDNA synthesis and thus allows single enzyme RT-PCR viral reverse transcriptase from avian murine virus (AMV RTase).
Asymmetric PCR: It is used to generate single-strand copies of a DNA sequence which can be directly used for DNA sequencing. The two primers (100 : 1 ratio) are such adjusted in the reaction mixture that one of them is exhausted about 10 or more cycles. After these cycles only a single strand of DNA segment is copied and these copies are the ideal starting materials for DNA sequencing. This variation is known as asymmetric PCR.
AP-PCR: Arbitrary primed PCR (AP-PCR) is a type of random amplified polymorphic DNA (RAPD) where single primers of 10 to 50 bases are used to amplify genomic DNA in PCR. Welsh and McCleland (1990) developed the arbitrary primed – PCR and carried out finger printing of genomes with arbitrary primers.
Polymerase chain reaction is useful in diverse areas of molecular biology, medicines and biotechnology.
- Diagnosis of pathogens: PCR is commonly used for diagnosis of infections caused by viruses (e.g. HIV-1, HIV-2, Herpes simplex virus, Hepatitis B virus etc.), bacteria (Mycobacterium tuberculosis, Helicobacter pylori, Mycoplasma pneumoniae etc), fungi (Candida albicans) and protozoa (Toxoplasma gondii, Trypanosoma cruzi etc).
- Diagnosis of plant pathogens: Various plant pathogens are detected by using PCR such as viruses (plum pox virus, cauliflower mosaic virus), fungi (Verticillium spp., Laccaria spp., Phytophthora spp. etc), mycoplasms bacteria (Agrobacterium tumifaciens, Rhizobium leguminosarum, Xanthomonas compestris etc) and nematodes (Meloidogyne incoginta) etc.
- Inherited diseases: Inherited disorders are caused by gene mutations passed on from parents to their children e.g. hemophilia, cystic fibrosis etc. PCR is used to amplify gene sequences which can then be screened for disease causing mutations.
- Research: PCR is used extensively as a research tool for identification of new species. Many bioactive microbial species are isolated from various extreme environment such as soil, water, air, sediments etc. DNA fingerprinting of new microorganisms is carried out to confirm their identity by comparing with the DNA sequences of known microorganisms.
- Cancer research: Polymerase chain reaction has been widely used in studies for the role of genes in cancer. Tumour-supressor genes and mutations in oncogenes have been identified in DNA from tumors using PCR-based strategies.
- Biotechnology: PCR has played major role in the production of recombinant proteins. Insulin and growth hormones are recombinant proteins, widely used as drugs and recombinant vaccines are developed for hepatitis B virus. It is an important tool in the biotechnology industries of research institutes.
- Forensic science: PCR is most applicable in forensic science where it is being used in search of criminals through DNA fingerprinting technology. PCR allows amplification of DNA from individual hairs, stains of blood or seminal fluid having partially degraded DNA. Analysis of variable sequences is also used in tissue typing to match organ donors with recipients.
- DNA polymorphism: PCR is used to study DNA polymorphism in the genome using known sequences as primers. PCR can be used to study RFLPS (restriction fragment length polymorphisms) as well as RAPDS (random amplified polymorphic DNA).
- Gene therapy: PCR proves to be immense help in monitoring a gene in gene therapy experiments. This PCR technology provides shortcuts for many cloning and sequencing applications.