The biosynthesis of a protein or a polypeptide in a living cell is called as translation. The genetic information stored in DNA is passed to RNA (transcription) and it expressed in the language of proteins (translation). Translation occurs by similar mechanisms of prokaryotes and eukaryotes and it is described in five stages.
- Activation of amino acids (Aminoacyl tRNA synthetases).
- Initiation (Binding of a ribosome to mRNA).
- Elongation (Repeated addition of amino acids).
- Termination and release (Release of new polypeptide chain).
- Folding and post-translational processing (polypeptide must fold into three- dimensional conformation and it may undergo enzymatic processing).
Translation requires the use of energy by the cl which is provided by the hydrolysis of GTP and ATP. Guanosine-triphosphate (GTP) is used for ribosome movement and in binding of accessory factors. Adenosine triphosphate (ATP) is used to change tRNAs and in removing secondary structure from mRNA. Protein synthesis requires many components such as amino acids, ribosomes, mRNA, tRNA, protein factors and energy sources (ATP and GTP).
Activation of Amino Acids:
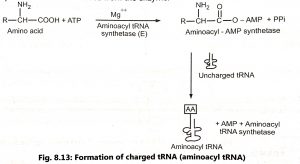
Cytoplasm contains 20 different amino acids and they are activated by a specific activating enzyme known as the aminoacyl synthetase and ATP before the attachment with its specific tRNA. The correct amino acid is attached to the tRNA by a type of enzyme called an aminoacyl-tRNA synthetase (aminoacylation/charging). The process of transfer of activated amino acids to tRNA is called charging of tRNA. The tRNAs are specific to theⅠr specific amino aminoacyl-tRNA synthase enzyme. Enzyme than catalyses a reaction in which the ATP loses two phosphates and is coupled to the amino acid as AMP to form aminoacyl AMP (Fig. 8.13). The tRNA molecule binds to the enzyme, which transfers the amino acid from the aminoacyl – AMP to the tRNA to form aminoacyl-tRNA. The aminoacyl-tRNA molecule produced is then reⅠeased from the enzyme.
Initiation:
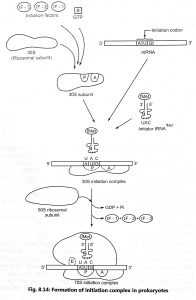
The first step in translation involves the binding of the small ribosomal subunit to the mRNA and use of specific initiating tRNA molecule. In prokaryotes, tRNA molecule is acylated with the modified amino acid (N-formyl methionine). Both tRNAfMet recognize the codon AUG but only tRNAfMet is used for initiation. The tRNAfMet molecule is first acylated with methionine and an enzyme adds a formyl group to the amino group of the methionine. In eukaryotes, the initiating tRNA molecule is charged with methionine but formylation does not occur.
The initiation of polypeptide synthesis in prokaryotes requires 30S and S0S ribosomal subunit, mRNA, tRNAfMet ,initiation factors(IF-1,IF-2 and IF-3),GTP and magnesium ions. Eukaryotic cells have at least nine initiation factors (eⅠF2, eⅠF23, eⅠF 3, eⅠF4A, eⅠF4B, eⅠF4E ,eⅠF4G, eⅠF5, eⅠF6). The initiation factors bind to 30S ribosomal subunit in the presence of GTP to form 30S-ⅠF complex. The 30S-ⅠF complex binds to the region of the mRNA with The AUG initiation codon. Each mRNA at its untranslational region consists of a ribosome binding site for every polypeptide in the form of polycistronic message. This ribosome binding site (5’- AGGAGGA-3′) is known as Shine-Dalgarno sequence which is important in the binding of mRNA to the 30S-ⅠF complex.
The ribosome has three important binding sites such as aminoacyl-tRNA binding site (A) peptide binding site (P) and exit site (E). The A site receives all the incoming charged tRNA.whereas the P site possesses the previous tRNA with the new polypeptides. The tRNAfMet directly binds with ‘P’ site. The E site is the site from which the ‘uncharged’ tRNAs leave during elongation. Factor IF-1 binds at the A site and prevents tRNA binding at this site during initiation.
The IF-2 which has combined with GTP, permits the initiator tRNA (tRNAfMet) to bind to the 30S ribosomal subunit (Fig. 8.14). A 30S ribosome unit is bound with 505 unit to produce 70S initiation complex. Similarly, in eukaryotes the 405 initiation complex is attached to 605 ribosomal subunit and forms the complete 805 initiation complex. The GTP bond to IF2 is hydrolyzed to GDP and Pi, which are released from the complex. All three initiation factors also depart from the ribosome. The initiation complex is now ready for elongation.
Elongation:
Elongation requires the initiation complex, aminoacyl-tRNAs, GTP and elongation factors. The addition of amino acids to the growing polypeptide chain as per codon on mRNA is called elongation of chain. Elongation of chain occur in three phases.
- Binding of aminoacyl- RNA.
- Peptide bond formation.
- Translocation.
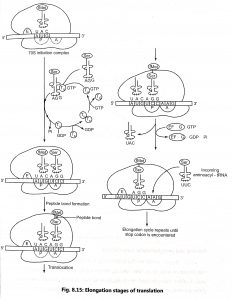
- Binding of aminoacyl-tRNA: The ribosome (70S) possess the tRNA in the P-site, whereas the A site is free to receive the next aminoacyl-tRNA according to the codons on MRNA. Aminoacyl-tRNA bind to the protein elongation factor EF-Tu and a molecule of GTP. GTP hydrolysis releases EF-Tu-GDP and EF-Tu is recycled. Second elongation factor (EF-Ts) binds to EF-Tu and displaces the GDP (Fig, 8.15). GTP binds to the EF-Tu-EF-Ts complex to produce EF-Tu-GTP complex by releasing EF-Ts. Aminoacyl-TRNA binds to the EF-Tu-GTP and that complex can bind to the A site in the ribosome.
- Peptide bond formation: A peptide bond is formed between the two amino acids bounded to their TRNAS to the A and P sites on the ribosome. First, the bond between the amino acid and the tRNA in the P site is breaked and form free fMet and its TRNA. The peptide bond is formed between the free fMet and the Ser attached to the tRNA in the A site (Fig. 8.15). The enzyme peptidyl transferase catalyses the formation of peptide bond.
- Translocation: The ribosome moves to the next codon of the mRNA (towards 3′-end) after the formation of peptide bond. This process is called translocation. Translocation requires the activity of another protein elongation factor, EF-G (In eukaryotes, eEF-2). An EF- G-GTP complex binds to the ribosome and GTP is hydrolyzed to supplies energy to move mRNA. Translocation of ribosome occurs along with displacement of the uncharged 1RNA away from the ‘P’ site.
Termination and Release:
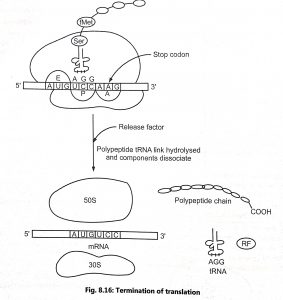
The polypeptide chain is continuously elongated until a termination codon on mRNA reaches to ribosome. The termination of translation is signaled by one of three stop cadons such as UAA, UAG and UGA. The ribosome recognize a stop codon with the help of proteins called termination factors or release factors (RF). In prokaryotes, there are three release factors (RF-1, RF-2, RF-3). RF-1 recognizes UAA and UAG and RF-2 recognizes UAA and UGA. The RF-3 activates the RF-1 and RF-2, hence, it is called ‘stimulatory(s) factor’. In eukaryotes,there is only one RF protein (eRF-1) which is active with codons UAA, UAG and UGA.
The specific termination events triggered by the release factors are release of the polypeptide from the tRNA in the P-site of the ribosome, release of the tRNA from the ribosome and the dissociation of the two ribosomal subunits and the RF from the mRNA.
Folding and post-translation processing:
After release, some of the processing events occur in the polypeptide chain. These modifications include protein folding, trimming by proteolytic degradation, intein splicing and covalent changes which are collectively known as post-translational modifications. Many different chemical modifications of the side chains of amino acids or the amino and carboxyl termini of proteins are found. Modifications may involve addition of small groups such as methylation, phosphorylation, acetylation and hydroxylation. Some modifications may occur by the addition of larger molecular structures such as lipid and oligosaccharides.
