Contents:
Transcription in Eukaryotes
Introduction
Transcription is a fundamental biological process that regulates gene expression in eukaryotic cells. Eukaryotic transcription is a multistep process that involves the conversion of DNA sequences into mRNA, which is then translated into proteins. This process is crucial for the development, growth, and function of living organisms.
Eukaryotic transcription is more complex than prokaryotic transcription because eukaryotic cells have a nucleus that separates the transcription and translation processes. Besides, eukaryotic cells have more regulatory mechanisms, and the RNA molecules undergo posttranscriptional modifications before they become functional
Enzymes Involved in Eukaryotic Transcription
Eukaryotic transcription involves the activity of a large number of enzymes and proteins. Some of the important enzymes involved are:
1. RNA polymerase
RNA polymerase is the main enzyme responsible for catalyzing the synthesis of RNA from DNA templates during transcription. Eukaryotic cells have three types of RNA polymerases: RNA polymerase I, II, and III. Each of these polymerases is an enzyme that catalyzes the synthesis of RNA from a DNA template during transcription.
2. Transcription factors
Transcription factors are proteins that bind to specific DNA sequences called the promoter region, which is located upstream of the gene to be transcribed. These factors recruit RNA polymerase and help in the initiation of transcription.
3. Histone-modifying enzymes
Histone proteins are involved in packaging DNA into a compact structure called chromatin. Histone-modifying enzymes add or remove chemical groups from histones, which can alter the chromatin structure and regulate gene expression.
4. Chromatin remodeling complexes
Chromatin remodeling complexes are protein complexes that can change the conformation of chromatin by moving, ejecting, or repositioning nucleosomes. This allows access to DNA by the transcription machinery and facilitates transcriptional regulation.
5. Coactivators and corepressors
Coactivators are proteins that enhance transcriptional activation, while corepressors are proteins that suppress transcriptional activation. They function by interacting with transcription factors and other proteins to modulate the transcriptional output.
Process of Eukaryotic Transcription
Step 1- Recognition
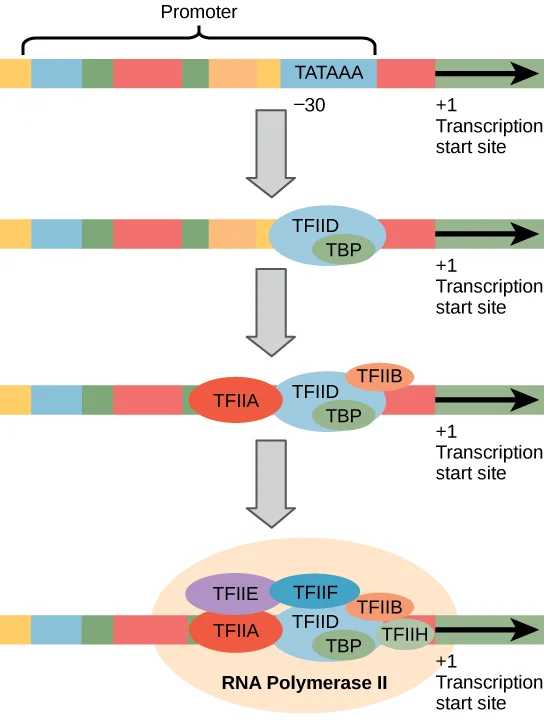
The first step in eukaryotic transcription is the recognition of the promoter region by the RNA polymerase enzyme. The promoter is a DNA sequence that is located upstream of the coding sequence. The promoter contains two important elements: the TATA box and the transcriptional start site (TSS). The TATA box is a conserved sequence of about 25 nucleotides, and it is recognized by a protein complex called the TATA-binding protein (TBP). TBP binds to the TATA box and helps to recruit other proteins that facilitate transcription initiation.
The TSS is the first nucleotide in the coding sequence, and it determines where the transcription starts. The TSS is usually preceded by a short sequence that is important for initiating transcription. The TATA box and the TSS are essential elements of the promoter, and they determine the efficiency and specificity of transcription initiation.
Step 2: Initiation
The initiation of transcription involves the assembly of a large protein complex called the preinitiation complex (PIC). The PIC is composed of various transcription factors and RNA polymerase II (Pol II). The assembly of the PIC is a multi-step process, and it involves the binding of transcription factors to the promoter region.
The general transcription factor TFIIA helps to recruit the TBP to the TATA box. Once TBP is bound to the TATA box, other transcription factors, including TFIIB, TFIIF, and TFIIH, join the complex. TFIIH is a kinase that phosphorylates the Pol II enzyme and helps to melt the DNA strands at the start site to create an open complex. The open complex is a stretch of unwound DNA that exposes the template strand for transcription.
Step 3: Elongation
After the formation of the open complex, the elongation phase of transcription begins. During elongation, RNA polymerase II synthesizes an RNA molecule that is complementary to the template strand of the DNA. The RNA molecule grows in length as RNAPII moves along the DNA template.
The elongation process is dynamic and productive, and the rate of elongation is influenced by many factors, including the sequence of the DNA template, the presence of nucleosome-bound DNA, and the availability of nucleotides. RNA polymerase II elongates the RNA molecule at a rate of about 20 to 30 nucleotides per second.
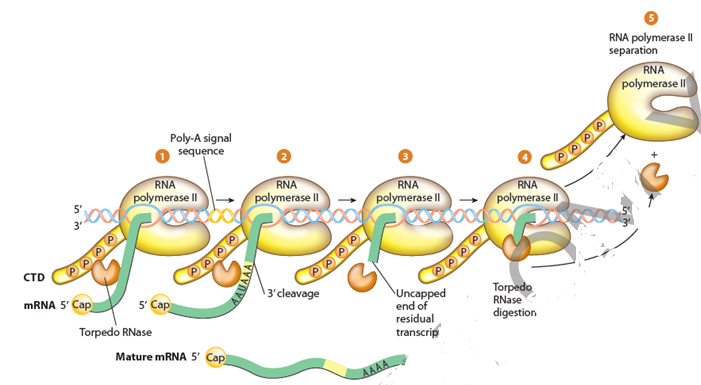
Step 4: Termination
The termination of transcription is the process by which RNA polymerase II dissociates from the DNA template. Termination can occur in two ways: through a specific signal sequence on the DNA or through a mechanism that involves the interaction of multiple factors.
The specific signal sequence on the DNA is called the polyadenylation signal. This sequence is located downstream of the coding sequence, and it signals the RNA polymerase to stop synthesizing RNA and release it from the DNA. The polyadenylation signal is recognized by a complex of proteins called cleavage and polyadenylation factors (CPF).
Alternatively, termination can occur through a more complex mechanism that involves the interaction of Pol II, elongation factors, and chromatin-modifying enzymes. This mechanism is regulated by various factors, including DNA topology, RNA secondary structure, and the activity of chromatin remodeling enzymes.
Step 5: Processing
After transcription, the newly synthesized RNA molecule undergoes several posttranscriptional modifications before it becomes functional. These modifications include capping, splicing, and polyadenylation.
The mRNA molecule is capped at the 5′ end with a modified guanine nucleotide that protects it from degradation and helps in translation. Splicing is the process of removing introns from the pre-mRNA and joining exons to form a functional mRNA molecule. Polyadenylation is the process by which a long poly(A) tail is added to the 3′ end of the mRNA molecule, which also helps in the stabilization, transport, and translation of the mRNA.
Conclusion
Eukaryotic transcription is a complex process that involves the recognition, initiation, elongation, termination, and processing of the newly synthesized RNA molecule. Each step of this process is regulated by various factors, including transcription factors, elongation factors, chromatin remodeling enzymes, and posttranscriptional modifiers.
Understanding the various steps involved in eukaryotic transcription is crucial for unraveling the underlying mechanisms that regulate gene expression. This knowledge can help in the development of new therapies that target various diseases that arise from aberrant transcription.
