Contents:
Tryptophan Operon
The tryptophan operon is a cluster of genes in bacteria that is responsible for the production of proteins involved in the biosynthesis of the amino acid tryptophan. This operon is regulated by a negative feedback mechanism, where the presence of tryptophan acts as a repressor to prevent further production of the amino acid synthesis of the amino acid tryptophan. The operon includes the trpE, trpD, trpC, trpB, and trpA genes, which encode for enzymes involved in the biosynthesis of tryptophan. The regulation of the operon is based on the presence or absence of tryptophan in the cell, which allows the bacteria to adapt to changes in their environment.
The tryptophan operon is one of the most well-studied genetic systems in bacteria and has played a fundamental role in the development of molecular biology. The operon has been extensively investigated in Escherichia coli (E. coli) and Salmonella typhimurium, two model organisms commonly used in molecular biology research.
The structure and function of the tryptophan operon in E. coli serve as an excellent example of the concept of gene regulation based on feedback inhibition. In this article, we will provide a comprehensive overview of the structure, regulation, and function of the tryptophan operon, focusing on E. coli.
Discovery of the Tryptophan Operon
The tryptophan operon was first discovered in E. coli by Jacques Monod and his colleagues in the early 1950s. At the time, Monod was working on the regulation of the bacterial enzyme β-galactosidase, which is involved in the metabolism of lactose. One of his graduate students, François Jacob, discovered that the gene for β-galactosidase was controlled by a system of regulatory genes that could respond to different environmental signals.
This discovery led Monod and Jacob to perform a series of experiments to try to understand how bacteria regulate the expression of their genes. One of their most famous experiments involved the use of E. coli strains that were deficient in tryptophan. They found that when these strains were grown in a medium that contained no tryptophan, they were unable to synthesize the amino acid and could not grow. However, when tryptophan was added to the medium, the bacteria were able to synthesize the amino acid and proliferate.
This led Monod and Jacob to hypothesize that the genes involved in tryptophan biosynthesis were regulated by a genetic system that responded to the availability of the amino acid in the environment. Further experiments confirmed that the tryptophan operon was indeed regulated by a negative feedback system that prevented transcription and translation of the genes involved in the biosynthesis pathway when tryptophan was present in the medium.
Structure of the Tryptophan Operon
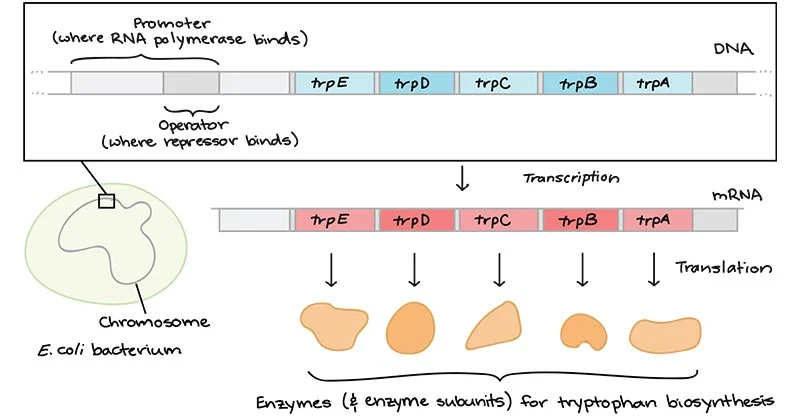
The tryptophan operon is made up of five genes, designated trpE, trpD, trpC, trpB, and trpA, that are involved in the biosynthesis of tryptophan. These genes are arranged in a linear fashion on the bacterial chromosome and are under the control of a single promoter and operator region that regulates their expression.
The operator region is located between the promoter and the first gene of the operon, trpE and it consists of two 21-base pair DNA sequences, known as operator I and operator II, which are separated by a 3-base pair spacer. These operator sequences bind to the regulatory protein, known as the trp repressor, which is encoded by the regulatory gene trpR located upstream of the promoter region.
When tryptophan is absent in the immediate environment of the bacterium, the trp repressor is unable to bind to the operator sequences and the RNA polymerase can initiate the transcription of the genes in the operon. This results in the production of all the enzymes involved in the tryptophan biosynthesis pathway, leading to the biosynthesis of the amino acid.
However, when tryptophan is present, it binds to the trp repressor protein, which then undergoes a conformational change that allows it to bind to the operator sequences. This binding prevents the RNA polymerase from initiating transcription and leads to the inhibition of the genes in the tryptophan operon. This negative feedback system ensures that the biosynthesis of tryptophan is regulated in response to the availability of the amino acid in the environment.
Regulation of Gene Expression
The expression of the tryptophan operon in E. coli is regulated by a mechanism called attenuation, which involves a combination of transcription and translation control. Attenuation is a process where the RNA polymerase initiates transcription of the operon, but the process is terminated before the operon is transcribed fully.
The regulation of the tryptophan operon is based on the presence or absence of tryptophan in the cell. In the presence of tryptophan, the expression of the operon is down-regulated because the cell already has enough tryptophan. Conversely, in the absence of tryptophan, the expression of the operon is up-regulated as the cell needs to synthesize more tryptophan.
The regulation of the tryptophan operon in E. coli is controlled by the trp repressor protein. The trp repressor protein is a tetrameric protein that binds to the operator sequence of the operon and prevents the RNA polymerase from transcribing the operon. The operator sequence is located downstream of the promoter region and upstream of the first gene (trpE) in the operon.
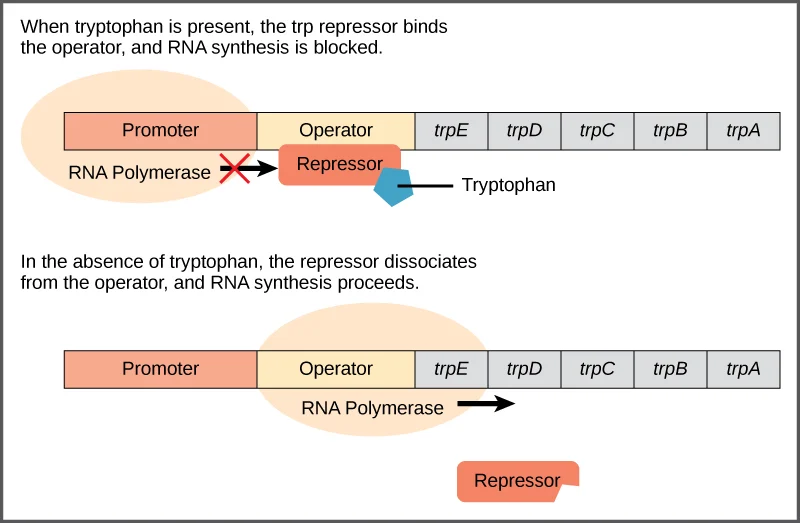
The trp repressor protein binds to the operator sequence only when it is bound to tryptophan. In the absence of tryptophan, the trp repressor protein undergoes a conformational change, making it unable to bind to the operator sequence. As a result, the RNA polymerase proceeds with the transcription of the operon.
Attenuation Mechanism
The attenuation mechanism is a regulatory process that allows the cell to control the gene expression of the tryptophan operon. Attenuation is based on the formation of different secondary structures in the polycistronic mRNA molecule that influence the transcription of the operon.
The attenuation mechanism is based on the formation of four different RNA secondary structures within the nascent mRNA molecule. These are called the leader sequences, which are located upstream of the first gene (trpE) in the operon.
The formation of the different RNA secondary structures depends on the availability of tryptophan in the cell. The secondary structures that form are different in the presence and absence of tryptophan, which in turn affect the ability of the RNA polymerase to transcribe the operon.
In the presence of tryptophan, the trp repressor protein binds to the operator sequence, which allows the RNA polymerase to start transcription. However, the presence of tryptophan affects the formation of the different secondary structures in the nascent mRNA molecule.
Specifically, attenuation is based on the formation of a stem-loop structure known as the trp leader. The trp leader can either form or not form depending on the availability of tryptophan. The formation of the trp leader depends on the interaction between two sequences in the nascent mRNA molecule called the leader peptide and the tryptophan codon.
The leader peptide sequence consists of 14 amino acids, which are encoded by the first 14 nucleotides of the mRNA molecule. The leader peptide has two tryptophan codons that can either bind to tryptophan-tRNA or remain unbound. The tryptophan-tRNA is the aminoacyl-tRNA molecule that interacts with the trp codon to add tryptophan to the growing polypeptide chain.
If tryptophan is abundant in the cell, the tryptophan-tRNA is readily available and binds to the tryptophan codon in the nascent mRNA molecule. This forms a stable structure, known as the terminator, which halts the transcription of the operon.
Alternatively, if tryptophan is not abundant, the tryptophan-tRNA is not readily available and cannot bind to the tryptophan codon. This results in the formation of a different secondary structure called the anti-terminator, which allows the RNA polymerase to continue the transcription of the operon.
The ability of the RNA polymerase to continue transcription of the operon is determined by the formation of the trp leader stem and the interaction between the leader peptide and the tryptophan codon. The anti-terminator structure folds only when the leader peptide interacts with the trp codon without the tryptophan-tRNA attached to it.
Applications of the Tryptophan Operon
The study of the tryptophan operon has been instrumental in the field of molecular biology and has led to important insights into the regulation of gene expression in prokaryotes. The operon was one of the first examples of negative gene regulation, and it helped to inspire the development of the regulatory gene theory, which proposes that many genes are regulated by regulatory proteins that bind to specific DNA sequences and control their expression.
The tryptophan operon has also been used as a tool in genetic engineering and synthetic biology. Researchers have developed plasmids that contain the tryptophan operon and can be used to control the expression of genes of interest in bacterial cells. These plasmids can be designed to respond to different small molecules, allowing researchers to control gene expression in response to changes in the environment or other stimuli.
Conclusion
The tryptophan operon is an excellent example of the regulation of gene expression based on feedback inhibition. The regulation of the operon in E. coli is based on the presence or absence of tryptophan in the cell.
The trp repressor protein controls the operon’s expression by binding to the operon’s operator sequence, thereby preventing the RNA polymerase from transcribing the operon. The attenuation mechanism is based on the formation of different RNA secondary structures in the nascent mRNA molecule, which regulates the transcription of the operon.
The formation of the different secondary structures in the nascent mRNA molecule is based on the interaction between the leader peptide and the tryptophan codon. The different secondary structures that form allow E. coli to adapt to changes in their environment and ensure that the cell is synthesizing the correct amount of tryptophan. The tryptophan operon remains an essential system in molecular biology and a subject of active research to this day.
References:
- https://en.wikipedia.org/wiki/Trp_operon
- https://www.medschoolcoach.com/operons-prokaryotic-gene-expression-part-1-mcat-biology
- https://www.medschoolcoach.com/operons-prokaryotic-gene-expression-part-1-mcat-biology
