Contents:
Bacterial Infection: Symptoms, Cause, and Diagnosis
INTRODUCTION- BACTERIAL INFECTIONS
Bacteria are microscopic, single-celled organisms. There are thousands of different kinds of bacteria that live in every conceivable environment all over the world. They live in soil, seawater, and deep within the earth’s crust. Some bacteria have been reported even to live even in radioactive wastes. Some bacteria live in the bodies of people and animals—on the skin and in the airways, mouth, and digestive and genitourinary tracts—often without causing any harm.
Bacterial infections occur when harmful bacteria enter our body or the existing bacteria get out of balance. The bacteria that cause disease are called pathogens. Sometimes bacteria that normally reside harmlessly in the body can also cause disease. Bacteria can cause disease by producing harmful substances like toxins or by invading the tissues.
Bacteria can be classified in several ways:
- Scientific names: Their scientific name is genus followed by species e.g. example, Clostridium botulinum. Within a species, there may be different types, called strains which differ in genetic makeup and chemical components. Certain drugs and vaccines are effective only against certain strains.
- Staining: Gram stain is the most commonly used stain for bacterial staining. Some bacteria stains blue. They are called gram-positive. Others stain pink. They are called gram-negative. Gram-positive and gram-negative bacteria stain differently because their cell walls are different. They also cause different types of infections, and different types of antibiotics are effective against them.
- Shapes: Bacteria may also be classified on the basis of three basic shapes: spheres (cocci), rods (bacilli), and spirals or helixes (spirochetes).
- Need for oxygen: Bacteria are also classified by whether they need oxygen to live and grow. The bacteria which need oxygen are called aerobes. Whereas the bacteria that have trouble living or growing when oxygen is present are called anaerobes. Some bacteria, called facultative bacteria, can live and grow both in the absence or presence of oxygen.
Most common causes of bacterial infections:
- Staphylococci—These often harmless bacteria commonly live in and on the body. Still some species can cause disease or infections.
- Streptococci—These are common bacteria. Some types can cause infections such as strep throat or other respiratory infections, including pneumonia.
- Haemophilus influenzae—These are also common type of bacteria that can sometimes cause infections. Harmful types can cause diseases that include respiratory infections, ear infections, etc.
- E Coli—These bacteria commonly live in the GI tract of animals and humans. Some can cause food poisoning if transmitted through improperly cooked food or other food products that have been contaminated.
- Pylori—These bacteria are a common cause of stomach ulcers.
- Salmonella—This is another food-borne pathogen that causes diarrhea or food poisoning. Some bacterial diseases can be identified clinically. However, most of the bacteria cause a wide range of clinical syndromes. And a single clinical syndrome may result from infection with any one of the many bacteria. Therefore, it is necessary to use microbiological laboratory methods to identify a specific etiologic agent. Diagnostic medical microbiology is the discipline that identifies etiologic agents of disease. The job of the clinical microbiology laboratory is to test specimens from patients for micro-organisms that are (or may be) a cause of illness and to provide information about the in vitro activity of antimicrobial drugs against the microorganisms identified. Infectious disease with bacterial etiology can be confirmed with the laboratory procedures summarized in Fig.1.
Considerable efforts have been made for the development of rapid, sensitive, and specific assays for the detection of causative organisms. In the last few years, many new techniques have entered into the field of the early diagnostic process of bacterial infections. Diagnosis of bacterial infections can be classified broadly into the following four types:
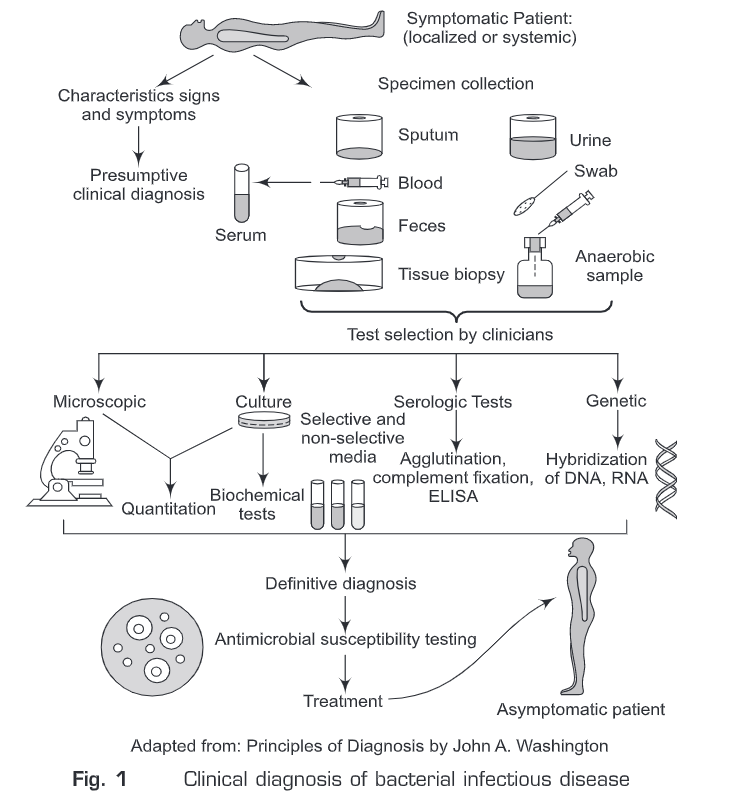
Microscopic Diagnosis of Bacterial Infection
- Unstained Method:
- It is commonly used to diagnose bacterial vaginosis. It includes wet preparation and potassium hydroxide (KOH) examination for microscopic examination of samples from the vaginal mucosa and skin surrounding the vaginal opening.
- Dark-ground illumination examination: Fresh samples can be prepared into slides and studied under dark field illumination. A newer method involves preparing slides from dried fluid smears and staining them with fluorescein for viewing under UV light. This method is replacing dark-field examination because the slides can be transported to professional laboratories.
- Stained method: Gram staining, Acid-fast staining
Culture Method
- Non-radiometric blood culture systems: One of the most important tasks performed by the clinical microbiology laboratory is the detection of bloodstream infections. Rapid bacterial identification and susceptibility testing use fluorescence-based technology. They are widely used because of the lower contamination risks, higher isolation rates and shorter incubation periods. These methods are both rapid and sensitive but still false-positive and false-negative results are high.
- The rapid evaluation of bacterial growth and antibiotic susceptibility in blood cultures by selected ion flow tube mass spectrometry (SIFT-MS): Selected ion flow tube mass spectrometry measures metabolic gases in the headspaces of blood culture bottles which helps in achieving faster bacterial diagnosis.
- In many cases, the cause of a bacterial infection is confirmed by isolating and culturing microorganism either in artificial media (either liquid (broth) or on solid (agar)) or in a living host. In some cases, we can take advantage differential media (e.g., eosin methylene blue or MacConkey agar) which are commonly used for the isolation of enteric bacilli.
- Culture media can also be made selective by addition of compounds such as antimicrobial agents that inhibit the indigenous flora while permitting growth of specific micro-organisms resistant to these inhibitors.
- For example, Neisseria gonorrhoeae can be isolated with Thayer-Martin medium. Thayer-Martin medium contains vancomycin which inhibits Gram-positive bacteria, colistin which inhibits most of the Gram-negative bacilli, trimethoprim-sulfamethoxazole which inhibits
- Proteus species and other species that are not inhibited by colistin and anisomycin which inhibits fungi. The pathogenic Neisseria species, N gonorrhoeae and N meningitidis, are ordinarily resistant to the concentrations of these antimicrobial agents in the medium.
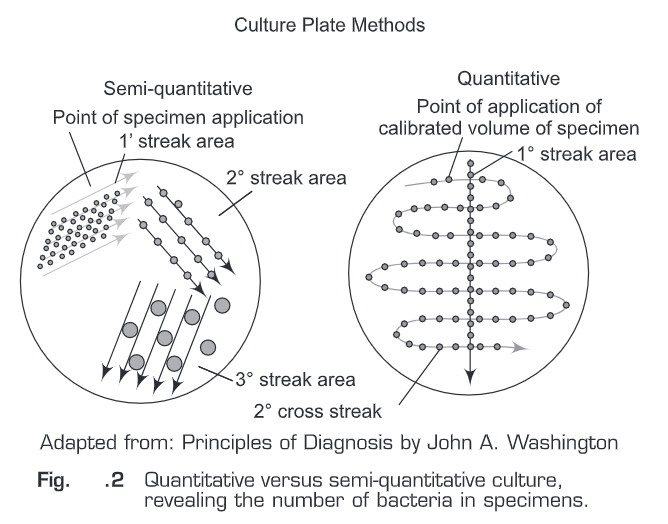
- Presence of bacterial infection can also be defined by the number of bacteria in specimens. For this quantitative cultures have to be performed.
- For other specimens a semi-quantitative streak method over the agar surface is sufficient. For quantitative cultures (Fig.2), a specific volume of specimen is spread over the agar surface and the number of colonies per milliliter is estimated.
- For semi-quantitative cultures, an unknown amount of specimen is applied onto the agar and then diluted by being streaked out from the inoculation site with a sterile bacteriologic loop.
- The growth on the agar is then reported semi-quantitatively as many, moderate, or few (3+, 2+, 1+ respectively), depending on how far colonies appear from the inoculum’s site. An organism that grows in all streaked areas is reported as 3+.
- Bacterial cultures are incubated at 35°C to 37°C in an atmosphere that consists of air, air supplemented with carbon dioxide (3-10%), reduced oxygen (micro-aerophilic conditions), or no oxygen (anaerobic conditions), depending upon the requirements of the micro-organism.
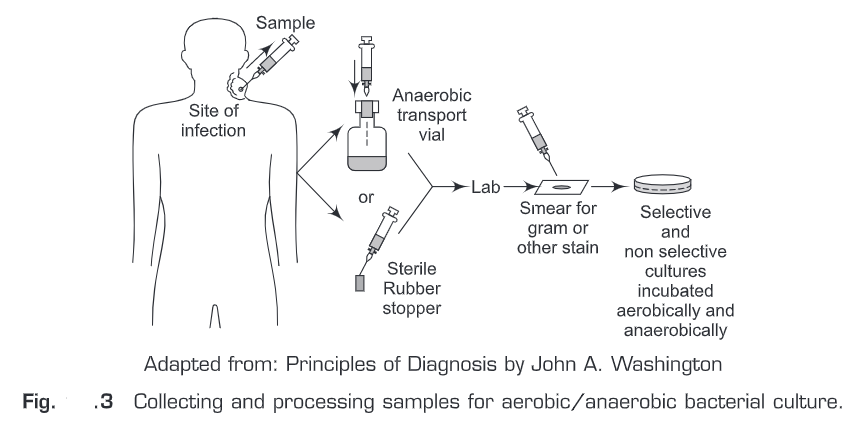
- Bacterial infections samples often contain aerobic, facultatively anaerobic, and anaerobic bacteria. Therefore, such samples are usually inoculated with general-purpose, differential, and selective media and finally are then incubated under aerobic and anaerobic conditions (Fig.3).
- The incubation period of cultures varies with the growth characteristics of the micro-organism. Most aerobic and anaerobic bacteria grow overnight, whereas some mycobacterium requires 6 to 8 weeks.
Antimicrobial Susceptibility
- The term susceptible means that the micro-organism is inhibited by an antimicrobial agent and implies that an infection caused by this micro-organism can be treated with the antimicrobial agent.
- In addition to microbial detection and isolation, the microbiology laboratory also determines the microbial susceptibility to antimicrobial agents.
- Many bacteria have unpredictable susceptibilities to antimicrobial agents which can be measured in vitro for the selection of the most appropriate antimicrobial agent.
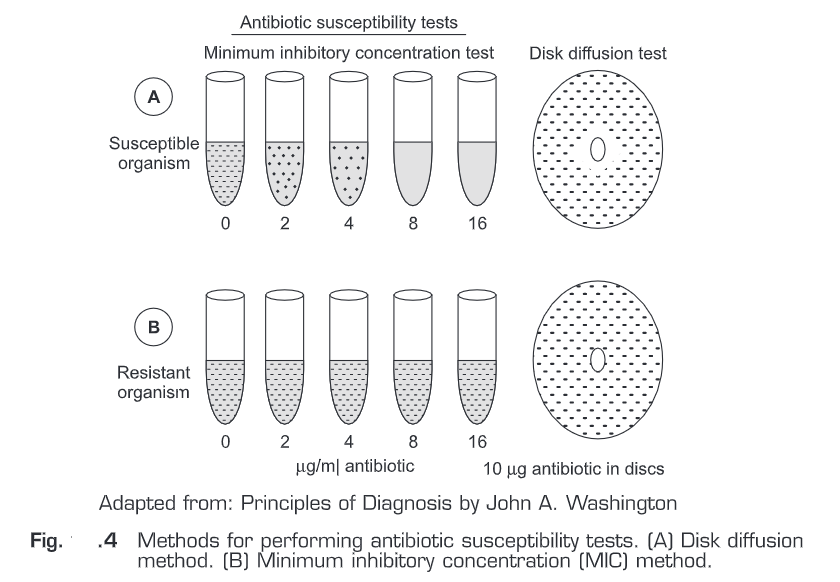
- Antimicrobial susceptibility tests are performed by either disk diffusion or a dilution method. In disk diffusion method, a suspension (standardized) of a particular micro-organism is inoculated onto an agar surface.
- Then paper disks containing various antimicrobial agents are applied on it. After overnight incubation, diameters of inhibition about the disks are measured and the results are reported as indicating susceptibility or resistance of the micro-organism to each antimicrobial agent tested.
- An alternative method is to dilute on a log2 scale each antimicrobial agent in broth to provide a range of concentrations and to inoculate each tube containing the antimicrobial] agent in broth with a standardized suspension of the micro-organism to be tested.
- The lowest concentration of antimicrobial agents that inhibits the growth of the micro-organism is the minimal inhibitory concentration (MIC).
- The MIC and the zone diameter of inhibition are inversely correlated. In other words, the more susceptible the micro-organism is to the antimicrobial agent, the lower the MIC and the larger the zone of inhibition.
- In Fig.4, two different micro-organisms are tested by both methods (disk diffusion or a dilution method) against the same antibiotic.
- The MIC of the antibiotic for the susceptible micro-organism is 8p1g/ml. The corresponding disk diffusion test shows a zone of inhibition surrounding the disk. In the second sample, a resistant micro-organism is not inhibited by the highest antibiotic concentration tested (MIC > 16 pg/ml) and there is no zone of inhibition surrounding the disk. The diameter of the zone of inhibition is inversely related to the MIC.
Molecular Methods
Molecular methods can be further be classified into two types:
- Direct methods— During the past decade, there has been unprecedented progress in molecular biology as well as in the application of nucleic acid technology to the study of the epidemiology of bacterial infection, some of which are summarized below.
- Polymerase Chain Reaction: The most widely used nucleic acid amplification and detection method, the Polymerase Chain Reaction (PCR) assay, has been found to have a substantial impact on the diagnosis of infectious diseases. Polymerase Chain Reaction amplifies minute amounts of specific microbial DNA sequences in a background mixture of host DNA and makes it a powerful diagnostic tool. The concept of DNA microarray hybridization has been introduced recently, and the technique has been applied in clinical diagnosis and many fields of research, e.g., functional genomics and genetic analysis. Microarrays are generally coupled with PCR where they serve as a set of parallel dot-blots to enhance products detection and identification. PCR assays that are currently available commercially for use in diagnostic laboratories include tests for the detection of C. pneumonia, Mycobacterium tuberculosis, Mycoplasma pneumoniae, etc.
- Nucleic Acid Hybridization and Restriction Fragment Length Polymorphism (RFLP) Analysis: In its simplest form, nucleic acid hybridization can be used for the detection of micro-organisms/specific resistance genes. It also confirms the results of microbial cultures or even detects organisms in clinical samples. In this method, a labeled synthetic nucleic acid probe detects the presence of target nucleic acids (oligonucleotide probe). The oligonucleotide probes directly identify microbial genetic sequences. Molecular hybridization has greater sensitivity and specificity than conventional techniques.
- RNA Typing or Ribotyping: RNA typing or ribotyping is a chromosomal detection technique. Genes that encode subunits of ribosomal RNA between species are conserved. The chromosomal DNA is digested by endonuclease which produces random fragment polymorphic patterns when probed with ribosomal RNA. In this method, E coli 165 ribosomal RNA acts as a probe for the endonuclease-digested chromosomal DNA. Because chromosomal nucleotide patterns usually vary from strain to strain but not within a strain, this technique can identify organisms and differentiate strains. The technique has application in the typing of Pseudomonas capacia, E coli, Salmonella typhi, etc. However, ribotyping is not very useful for gram-positive organisms.
- PCR Ribotyping: PCR ribotyping is the analysis of banding patterns obtained by gel electrophoresis of PCR amplified fragments of the 16S to 23S ribosomal RNA intergenic spacer regions. It has considerable advantages in terms of speed and technical ease and is useful in detecting and typing C difficile strains.
- Pulsed-Field Gel Electrophoresis: This is a widely used technique for analyzing a large amount of chromosomal DNA found in large bacterial chromosomal fragments generated by endonuclease digestion.
- Clamped Homogeneous Field Electrophoresis: This method is used to compare large chromosomal fragments generated by restriction endonuclease digestion. This technique is an easy way to compare isolates of a species. Clamped homogeneous field electrophoresis helps in the analysis of vancomycin-resistant enterococci. No reliable typing system previously has been able to identify strains of this organism beyond the species level.
- Multilocus Sequence Typing: This is a genome-based version of the conventional method of multilocus enzyme electrophoresis. It helps in the typing of various bacterial species by identifying DNA alleles from various organisms. This method involves PCR amplification and the nucleic acid sequencing of multiple internal fragments of housekeeping genes. The advantages of this method are that the culturing of pathogenic micro-organisms is avoided and that the sequencing data are unambiguous, easy to standardize, and electronically portable.
- Fluorescence-Based Amplified Fragment Length Polymorphism: This is a novel assay based on the fluorescent analysis of an amplified subset of restriction fragments. The fluorescence-based amplified fragment length polymorphism assay involves the selective PCR amplification of restriction fragments from a total digest of genomic DNA. It has been useful in the study of vancomycin-resistant enterococci.
- DNA Sequencing and Molecular Evolutionary Analysis: The sequencing of DNA refers to the enumeration of individual nucleotide base pairs along a linear segment of DNA. Single-stranded or double-stranded DNA generated by PCR can be used directly for DNA sequencing. Highly sensitive molecular techniques are capable of detecting single base-pair substitutions and resolving the mechanism of underlying complex variation and epidemiology.
- Indirect methods—The quest for surrogate biomarkers to define systemic inflammatory response syndrome (SIRS) has identified several potential candidates. For example, pro-calcitonin, C-reactive protein (CRP), tumor necrosis factor-alpha (TNF-α), and several interleukins appeared promising in initial studies. Several of them are described below.
- Cytokines: When a bacterium enters our body, it is confronted with two lines of defense: a humoral line and a cellular line. There are marked differences in the responses to gram-positive and gram-negative bacteria. Gram-negative bacteria contain lipopolysaccharide (LPS) as their major pathogenic determinant, whereas gram-positive bacteria contain a number of immunogenic cell wall components besides the highly deleterious exotoxins. The immunological response to gram-negative bacteria mainly involves leukocytes and the production of cytokines. For example, tumor necrosis factor-alpha (TNF-α), interleukin-1 (IL-1), and IL-6.
- Procalcitonin (PCT): PCT is a new marker that has been associated with inflammation and sepsis. It is a 116-amino-acid protein that is the precursor to calcitonin, The PCT plasma level in healthy individuals is low, generally below 0.1ng/ml. It has been found to be a more reliable marker in the diagnosis of sepsis than other measures.
- C-reactive protein (CRP): CRP, an acute-phase protein is released from the liver in response to pro-inflammatory cytokines and recruits monocytes in early infections. Serial CRP levels are useful in the diagnostic evaluation of neonates with suspected infection. Two CRP levels <1 mg/dL obtained 24-hour apart, 8 to 48 hours after presentation, indicate that bacterial infection is unlikely. The positive predictive value of elevated CRP levels is low, especially for culture-proven early-onset infections.
- Cell surface receptors: Quantitation of neutrophil (CD64) expression provides improved diagnostic detection of infection/sepsis as compared with the standard diagnostic tests used in current medical practice with a sensitivity of 87.9% and specificity of 71.2%. One more cell surface receptor CD11 is also found to be upregulated during infection.