The genome of about 50% of bacterial species and up to 90% of archaebacteria harbors the unique genetic region responsible for so-called “adaptive immune system” of bacterial cells. For instance, this ability is essential for bacteria, which are the threatening pathogens of humans, e.g. M. tuberculosis, Y. pestis, or S. pyogenes. The CRISPR–cas system is an adaptive immune system of bacteria and archaea, which protects the bacteria from invaders, including bacteriophages or phages and mobile genetic elements (MGEs). The CRISPR locus is present in 84% of archaea and 45% of bacteria. It was observed first in E.coli in K-12 chromosome. These element were also observed in chromosomes of Shigella dysenteriae and Salmonella enterica serovar Typhimurium.
Genes and genetic components of adaptive immune system of bacteria
- The genes and genetic components of this mechanism control the bacterial cell ‘s acquired protective reactions to the invaded foreign bacteriophage or plasmid nucleic acid.
- In addition, in the bacterial genome, genetic material from invaded DNA is memorised and becomes heritable. As a consequence, the next entry of the same foregn nucleic acid leads to the activation of particular defence reactions mediated by RNA that degrade invaded bacteriophageal or plasmid exogenous nucleic acid. The bacterial cell becomes “immune” to its particular pathogen in this manner.
- It has long been determined that the minimal amount of cells persists after penetration with a certain bacterial species with a particular bacteriophage and provides defence against this phage, thereby creating a new resistant bacterial population.
- Specific genetic elements known as CRISPR cassettes comprise the genome of these bacteria. They are found inside bacterial nucleoids in most cases.
CRISPR
- The acronym “CRISPR” is deciphered as a “clustered short palindromic repeat periodically interspaced.” This suggests the CRISPR cassette’s genetic structure, which contains genetic spacers of comparable length but distinct sequences of DNA interspersed with virtually identical direct DNA repeats.
- It was known to be novel defense system.
- There could be more than 100 spacers on a single CRISPR cassette.
- Moreover, the DNA sequence of a significant proportion of spacers seemed to be very similar to the DNA fragments of bacterial pathogens, such as bacteriophages.
- Multimeric enzymes with metal-dependent nuclease and integrase operations are Cas proteins.
Mechanism of CRISPR in bacteria
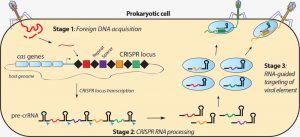
- CRISPR arrays remember” the virus or attacking apecies. The repeated attack makes the bacteria produce RNA segments from the CRISPR arrays to target the viruses’ DNA. The bacteria then uses Cas9 or a similar enzyme to cut the DNA apart disabling the virus.
- The mechanism of a new spacer acquisition starts when the bacteriophage releases its DNA into the bacterial cell cytoplasm.
- It consists of Cas1 / Cas2 protein complex hydrolysis of viral DNA accompanied by the release from the phage ‘s DNA of the so-called protospacer sequence and its further incorporation into the bacterial genome region of CRISPR. Both these activities contribute to the bacterial cell acquisition of the new spacer, which harbours unique international bacteriophageal DNA.
- Similarly , it is possible to inject new spacers from the same phage around the first phage. The bacterial cell therefore remains immune to the next infection that this phage induces.
- It activates the transcription of the main part of the CRISPR locus as another episode of phage infection occurs. This results in the synthesis of the primary long RNA transcript that is further cleaved by CRISPR-RNA (crRNA)-forming Cas proteins. To the phage sequence, this crRNA is complementary.
- At the next level, crRNA accounts for the nuclease operation of the Cas protein complex and delivers it to phage nucleic acid (‘guide RNA’). CrRNA recognises unique phage DNA sequence here and binds to it to cause the invaded nucleic acid to be killed by Cas nuclease.
- In addition to foreign DNA, unique Cas proteins can also kill RNA phages and phage messenger RNAs, depending on their form.
- These genetic adaptive immune pathways shield bacteria from phage infections as well as from the introduction by natural mutation or conjugation of unwanted genetic content.
- Specially engineered CRISPR-Cas systems (e.g., CRISPR / Cas9) have been engineered as the most effective genetic engineering methods that allow both prokaryotic and eukaryotic genomes to be reliably and conveniently edited.
Reference