Contents:
Electron Transport Chain
The electron transport chain is a sequence of four protein complexes that incorporate redox reactions to create an electrochemical gradient in a complete mechanism called oxidative phosphorylation that contributes to the formation of ATP. It occurs in both cellular respiration and photosynthesis in mitochondria.
The oxidation of glucose carbon atoms is carried out in glycolysis and the citric acid cycle, and the produced protons and electrons are stored in NADH and FADH2 molecules. The electrons from NADH and FADH2 are injected into the electron-transport chain in the inner membrane of the mitochondrion. When the electron passes through the electron-transport chain into the O2 reduction chamber (a protein complex), the protons in the mitochondrial matrix are expelled to the intermembrane space so as to generate the pH gradient between the matrix and the intermembrane space of a mitochondrion. The free energy stored in the resulting pH gradient drives the synthesis of ATP from ADP and Pi. This process is called oxidative phosphorylation.
Oxidation of NADH by O2 (2e– transfer) under standard biochemical condition releases 218 kJ of free energy, and an ATP synthesis (ADP + Pi → ATP) requires 30.5 kJ/mol free energy. Thus, several moles of ATP can be synthesized from 218 kJ of free energy of (½O2 + NADH →NAD+ + H2O) reaction. Actually, 218 kJ free energy is broken up into three packets. Each of the packets is coupled with one ATP synthesis. Peter Mitchell (1961) suggested the Chemiosmotic Hypothesis to explain oxidative phosphorylation and the electron transport chain. According to this principle, in order to create an electrochemical H+ gradient through the inner mitochondrial membrane, the free energy of electron transfer is maintained by pumping H+ (proton) from the mitochondrial matrix into the intermembrane vacuum. This gradient’s electrochemical potential is harnessed to synthesize ATP.
Sequence of electron transport
1. Complex I (NADH-Coenzyme Q reductase)
NADH electrons are inserted into Complex I (NADH-Coenzyme Q reductase), which is a protein complex with transmembrane multisubunit. In mitochondria, Complex I is possibly the largest protein component. NADH is donated by glycolysis, and the cycle of citric acid is oxidized here, passing 2 electrons to FMN from NADH. Then they are moved to the Fe-S clusters and eventually to coenzyme Q from Fe-S. During this process, 4 hydrogen ions, contributing to the electrochemical gradient, move from the mitochondrial matrix to the intermembrane space.
(NADH + H+) + CoQ + 4 H+(matrix) -> NAD+ + CoQH2 + 4 H+(intermembrane)
2.Complex II (Succinate- Coenzyme Q Reductase)
The electrons of FADH2 are transferred into Complex II (Succinate- Coenzyme Q Reductase). Complex II contains succinate dehydrogenase, three hydrophobic subunits,s and also contains covalently bound FAD, one [4Fe–4S], one [2Fe–2S], one cytochrome b560. 2 electrons are accepted by FAD within complex II as succinate oxidizes to fumarate. They are transferred to Fe-S clusters by FAD and then to coenzyme Q,
-
Succinate + FAD -> Fumarate + 2 H+(matrix) + FADH2
-
FADH2 + CoQ -> FAD + CoQH2
Co Enzyme Q- Coenzyme Q is made up of quinone and a hydrophobic tail, also known as ubiquinone (CoQ). Its aim is to act and pass electrons to complex III as an electron carrier.
3.Complex III (Coenzyme Q-Cytochrome c Reductase)
Electrons are then carried by a CoQ (coenzyme Q (ubiquinone)) to the Complex III (Coenzyme Q-Cytochrome c Reductase). It passes electron from reduced CoQ to cytochrome c. Complex III contains two b-cytochromes (bL (566 nm) and bH (562 nm)), one cytochrome c1, and one[2Fe–2S] cluster. The electrons are further transferred to Complex IV (Cytochrome c oxidase) which is an O2 reduction chamber. A cytochrome is a protein that comprises a group of hemes that is involved in electron transfer. During electron transfer, the heme groups alternate between ferrous (Fe2 +) and ferric (Fe3 +) states.
2 CoQH2(site 1) + CoQ(site 2) + 2 Cyt c(ox) + 2 H+(matrix) -> 2 CoQ(site 1) + CoQH2(site 2) + 2 Cyt c(red) + 4 H+(intermembrane)
4.Complex IV (Cytochrome c oxidase)
It catalyzes the oxidation of one electron by four consecutive reduced molecules of cytochrome c and the concomitant four-electron reduction of one molecule of O2. In addition to the heme and copper groups in complex IV, the cytochrome proteins a and a3 pass the donated electrons to the bound dioxygen species, turning them into water molecules.
2 cytochrome c(red) + ½O2 + 4 H+(matrix) -> 2 cytochrome c(ox) + 1 H2O + 2 H+(intermembrane)
When the electron passes through each Complex (Complex I, III, and IV), an amount of H+ is pumped out from the mitochondrial matrix to the inner membrane space, which can produce one mole of ATP. The electrons of NADH are injected into Complex I The received electrons are then passed to CoQ. FADH2 electrons are passed into Complex II, passing electrons from succinate to CoQ as well. The electrons then pass via complex III and complex IV and, through ATPase, build electrical potential and proton motive force and create ATP. ΔG of electron transport is conserved by pumping H+ from the mitochondrial matrix to intermembrane space, creating an electrochemical H+ gradient across the inner membrane, which drives ATP synthesis.
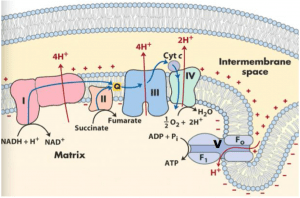
The respiratory chain is arranged in three loops corresponding to the three coupling sites. By a special vectorial arrangement of the electron-carrying molecules, an H+ adsorbing reaction occurs on the inside surface of the inner mitochondrial membrane (i.e., the surface facing the matrix). A concomitant H+-releasing reaction occurs on the outside surface of that membrane. As a result, an H+ gradient develops, with a higher concentration of H+ on the outside of the inner membrane. A reversible ATPase complex on the mitochondrial inner membrane is located in a region impermeable to water but accessible to OH- from. one side of the inner membrane and accessible to H+ from the other side. Thus ATP hydrolysis would be reversibly coupled to the translocation of OH- ions across the system with a stoichiometry of one OH- translocated per ATP molecule hydrolyzed. The proton gradient provides energy for the synthesis of ATP. Protein complexes (I, III and IV) catalyze the following reactions:
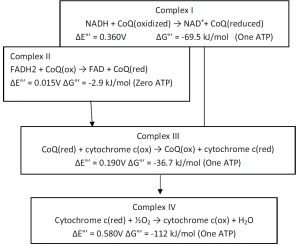
Mechanism of ATP synthesis:
The proton translocating ATP synthase (proton pumping ATPase, F1F0-ATPase) synthesizes ATP. It is found in the membrane of the inner mitochondria. Proton-translocating ATP synthase is a multi-subunit transmembrane protein. It has two major substructures, i.e., F1 and F0. F1 is a water-soluble peripheral membrane protein composed of 9 subunits (α3β3γδε), and F0 is a water-insoluble transmembrane protein composed of 10-12 subunits. There is a stalk between F1 and F0 containing oligomycin-sensitivity-cofactorring protein (OSCP) and coupling factor 6 (F6). Catalytic sites on β subunits assume different conformation per 120o turn. One of them binds ADP and Pi, and another catalyzes the attachment of Pi to ADP and extracts ATP from one of them.
