Contents:
Bacterial resistance to antibiotics
Resistance to glycopeptide antibiotics
- Vancomycin (isolated from the soil) and teicoplanin are the two most known glycopeptides and new semisynthetic glycopeptides dalbavancin, oritavancin, and telavancin are used clinically.
- They are mostly active against Gram-positive bacteria and bind to the terminal Dalanyl-D-alanine side-chains of peptidoglycan and prevent cross-linking in the cell wall.
- The outer membrane acts as the restriction for these antibiotics to act against the gram-negative bacteria.
- The resistance to these antibiotics was first noticed in vancomycin among enterococci mostly in E.faecium and E.faecalis and Staphylococcus aureus.
- The glycopeptide antibiotics are tetracyclic compounds with a core made up of seven amino acids (heptapeptide) to which two sugar moieties are bounded.
Vancomycin
Mode of action of Vancomycin
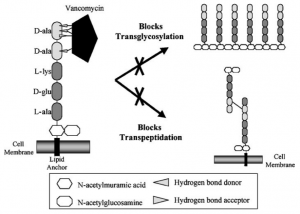
- Vancomycin inhibits cell wall synthesis by inhibiting peptidoglycan(cell wall) precursors forming a stable, noncovalent complex with the C-terminal D-Ala-D-Ala of murein monomers.
- These monomers D-Ala-D-Ala terminus of the peptidoglycan pentapeptide linker are held by five hydrogen bonds.
- This leads to the inhibition of the cross-linking in the peptidoglycan thus inhibiting the cell wall synthesis and simultaneously it causes a conformational change that blocks glycosyltransferase enzyme.
- This results in the inhibition of the transglycosylation, thus stopping peptidoglycan growth by blocking the addition of N-acetylmuramic and acid and N-acetylglucosamine in the growing chain.
Bactericidal effect of vancomycin on gram-positive bacteria
Resistance to Vancomycin
- Frequent use of oral vancomycin for Clostridium difficile infections and vancomycin for MRSA, enterococci including E. faecium strains and E. faecalis soon developed resistance to vancomycin and teicoplanin.
- Vancomycin-resistant enterococci (VRE) is the cause for more than 20% of all enterococcal infections.
- The resistance against these antibiotics occurred when the bacterium acquired and expressed operons that substitute a terminal D-lactate (D-Lac) or D-serine (D-Ser) for the terminal D-Ala in peptidoglycan precursors.
- This replacement results in a 1000-fold decrease in vancomycin affinity because of the loss of one of the five hydrogen bonds which used to hold the D-Ala-D-Ala of murein monomers (peptidoglycan precursors)
- Seven operons or gene clusters that have been reported(vanA, vanB, vanC, vanD, vanE, vanG and vanL) .
- Enterococci with the vanA gene confer to high resistance to both vancomycin and teicoplanin
- In contrast, vanB enterococci retain teicoplanin susceptibility and are resistant only to vancomycin
- VanC resistance is intrinsic and chromosomally encoded in some enterococcal species such as E.
gallinarum and the peptidoglycan precursor is D-Ala-D-Ser. - VanD and VanE resistance, but both are acquired. VanD uses D-Ala-D-Lac and VanE uses D-Ala-D-Ser.
MRSA and reduced glycopeptide susceptibility
- Enterococci are intrinsically resistant to multiple antibiotic classes and are able to acquire additional resistance mechanisms which make them reservoirs of antimicrobial resistance genes.
- MRSA or Methyl Resistant Staphylococcus aureus is resistant by VanA-type resistance.
- It is also caused by the resistance operons.
Aminoglycoside antibiotics
- Aminoglycoside (AG) antibiotics are used to treat many Gram-negative and some Gram-positive infections and, multidrug-resistant tuberculosis.
- Resistance to AGs arises through a variety of intrinsic and acquired mechanisms.
- AGs have been examined as potential treatments for fungal infections, Leishmaniasis parasitic infections, and for genetic diseases arising from premature termination codons, such as cystic fibrosis, Rett syndrome, and Duchenne muscular dystrophy
- AGs exert their antibacterial action by binding to the bacterial ribosome and interfering with bacterial protein translation.
Resistance to aminoglycoside antibiotics
- The aminoglycosides are hydrophilic sugars possessing a number of amino and hydroxy substituents.
- The bacterial ribosome consists of 70S subunits ie 50S and 30S subunits and the protein translation occurs at 3 sites – E, P, A sites
- The amine groups of AG are protonated at biological pH and it has a high affinity for nucleic acids, particularly the acceptor (A) site of 16S ribosomal RNA.
- Aminoglycoside binding to the A site interferes with the accurate recognition of cognate tRNA by rRNA during translation and may also disturb the translocation of the tRNA from the A site to the peptidyl-tRNA site (P site).
- Also, the AG binding site may be modified enzymatically by 16S ribosomal RNA methyltransferases. Ex- actinomycetes
- The bacteria that is infected by this antibiotic may resist themselves by producing RMTases to methylate their own 16S rRNA.
- The most common mechanism for aminoglycoside resistance is the chemical modification by enzymes called aminoglycoside-modifying enzymes (AMEs) which compromises their ability to interact with rRNA.
- There are three classes of these enzymes: aminoglycoside phosphatases (APHs),
aminoglycoside nucleotidyltransferases (ANTs) and aminoglycoside acetyltransferases (AACs). - Each AME modifies an AG at a specific position leading to some structural modifications.
Tetracycline antibiotics
- Tetracycline antibiotics are effective against Gram-positive and -negative bacteria, spirochetes, obligate intracellular bacteria, as well as protozoan parasites.
- Tetracyclines bind to bacterial ribosomes and interact with 16S ribosomal RNA (rRNA) target in the 30S ribosomal subunit, inhibiting the translation by interfering with the docking of aminoacyl-transfer RNA (tRNA) during elongation
- Chlortetracycline and oxytetracycline are clinically used tetracycline antibiotics.
Resistance to tetracycline antibiotics
- But resistance has been observed in many species including Escherichia coli, Klebsiella species, methicillin-resistant Staphylococcus aureus (MRSA), and Streptococcus pneumoniae
- The resistance was marked because of efflux, ribosomal protection, and enzymatic inactivation of tetracycline drugs.
- Most bacteria have multiple rRNA copies which confer target based mutation having low rRNA gene copy numbers leading to resistance of antibiotics.
- Unlike rRNA genes, genes encoding ribosomal proteins are single copy and mutations in these genes can confer antibiotic resistance.
- The most common tetracycline-specific efflux pumps are members of the major facilitator superfamily (MFS) of transporters.
- These proteins (MFS) exchange a proton for a tetracycline–cation (usually Mg2+) complex,
reducing the intracellular drug concentration and protecting the target ribosomes in the cell. - Drug binding alters the conformation of the repressor which do not allow to bind the DNA operator region and block transcription and are applicable to all Gram-negative efflux systems.
Fluoroquinolone antibiotics
- Quinolones have been a widely used class of synthetic antimicrobials used in both human and veterinary medicine.
- Quinolones are broad-spectrum antibiotics that are active against both Gram-positive and Gram-negative bacteria, including mycobacteria, and anaerobes.
- Chloroquine, one of the antibiotics are used in the treatment of urinary tract infections
- The fluoroquinolones selectively inhibit bacterial nucleic acid synthesis by disruptring the enzyme topoisomerases II and IV and DNA gyrase
Resistance to fluoroquinolone antibiotics
- Fluoroquinolones bind and inhibit two bacterial topoisomerase enzymes: DNA gyrase (topoisomerase II) which is required for DNA supercoiling, and topoisomerase IV which is required for strand separation during cell division.
- DNA gyrase consists of 2 GyrA and 2 GyrB subunits and topoisomerase IV composed of 2 ParC and 2 ParE subunits.
- DNA gyrase targets Gram-negative bacteria, whereas both topoisomerases are inhibited in
Gram-positive bacteria. - Mutations in gyrA, particularly involving the substitution of a hydroxyl group with a bulky hydrophobic group which leads to conformational changes inhibiting fluoroquinolone to bind.
- Alterations involving Ser80 and Glu84 of S. aureus grlA and Ser79 and Asp83 of S. pneumoniae
parC has led to quinolone resistance. - Topoisomerases are located in the cytoplasm and thus fluoroquinolones must cross the cell envelope to reach their target which provides resistance in Gram-negative bacteria.
- Efflux mechanism tends to work for a low level of Grampositive and Gram-negative bacteria.
- NorA expression confers resistance to hydrophilic quinolones, such as norfloxacin and ciprofloxacin, whereas NorB and NorC expression each confers resistance to hydrophilic quinolones and hydrophobic quinolones, such as sparfloxacin and moxifloxacin.
- Fluoroquinolones are used for treating Mycobacterium avium and multidrug-resistant M. tuberculosis and efflux-mediated resistance has been identified.
- A number of efflux pumps have been identified among Gram-negative bacteria, including AcrA in E. coli, which is regulated in part by the multiple antibiotic resistance
Chloramphenicol
- Chloramphenicol is a broad-spectrum antibiotic against several gram-positive and gram-negative bacteria, spirochetes, and Rickettsiae.
- It inhibits mRNA translation by binding to the 70S ribosomes of prokaryotes but does not affect 80S eukaryotic ribosomes.
- Chloramphenicol is lipid-soluble which let them diffuse through the bacterial cell membrane and reversibly binds to the bacterial 50S ribosomal subunit.
- The binding interferes with peptidyl transferase activity preventing the transfer of amino acids to the growing peptide chains and blocks peptide bond formation which blocks bacterial protein synthesis.
Resistance to chloramphenicol
- Chloramphenicol resistance(Cmr) was plasmid-borne and the resistance is due to the presence of an enzyme, chloramphenicol acetyltransferase (CAT), which catalyzes the acetyl-CoA dependent acetylation of the antibiotic at the C-3 hydroxyl group and sometimes by chloramphenicol phosphotransferases
- Chloramphenicol acetyltransferases (CATs) inactivates chloramphenicol. Cmr may also be due to the efflux of chloramphenicol via specific membrane-associated transporters.
- Cmr may also occur from mutations that reduce the permeability of outer membrane proteins, mutations in the 23S rRNA or target site modification by a 23S rRNA methylase.
Oxazolidinone antibiotics
- Linezolid is the first oxazolidinone available.
- It is involved in the treatment of complicated skin infections caused by methicillin-resistant S.
(MRSA), nosocomial pneumonia caused by MRSA, concurrent bacteremia associated with vancomycin-resistant Enterococcus faecium, and concurrent bacteremia associated with community-acquired pneumonia caused by penicillin-resistant S. pneumoniae. - Oxazolidinones bind to the 50S ribosomal subunit and they have no affinity to the 30S subunit
Resistance to the oxazolidinone antibiotics
- The decreased affinity of the ribosome for the oxazolidinones due to mutations in the central loop of domain V of the component 23S rRNA is the cause of resistance to these drugs
- Ribosomal resistance to oxazolidinones was shown in oxazolidinone resistant mutants by ribosomal binding and in vitro translation studies
Macrolide, lincosamide, and streptogramin (MLS) antibiotics
- These members of the MLS group of antibiotics inhibit bacterial protein synthesis by binding to a target site on the ribosome.
- Gram-negative bacteria are resistant due to the permeability barrier of the outer membrane.
- Staphylococcus aureus (S. aureus) is considered as the pathogen obtaining resistance to certain antibiotics.
Resistance to macrolide, lincosamide, and streptogramin (MLS) antibiotics
- Three mechanisms are responsible for acquiring resistance to MLS antibiotics in staphylococci: (1) target site modifications by methylation or mutation; (2) active efflux of antibiotics; or (3) inactivation of antibiotics.
- Target site modifications by methylation or mutation include target site modifications by a methylase encoded by one or more of the erm genes, methylating 23S rRNA and thereby altering binding sites for MLS antibiotics
- Active efflux of antibiotics involves a macrolide efflux pump is encoded by msr(A) and/ or msr(B) genes
- A third resistance mechanism, involving ribosomal mutation, has been reported in a small
a number of clinical isolates of S. pneumoniae.
Trimethoprim
- Trimethoprim is used for the treatment of enteric, respiratory, skin, and urinary tract infections
- It acts bacteriostatically by inhibition of dihydrofolate reductase (DHFR).
Resistance to trimethoprim
- The bacterial resistance to trimethoprim is mediated by
(i) a permeability barrier
(ii) a naturally insensitive intrinsic dihydrofolate reductase (DHFR)
(iii) spontaneous mutations in the intrinsic DHFR
(iv) increased production of the sensitive target enzyme by upregulation of gene expression or gene duplication
(v) horizontal acquisition (plasmid-mediated or conjugation) of dfr genes that encode resistant DHFRs.
Mupirocin
- The drug is widely used to treat Staphylococcus aureus infections and decolonization of methicillin-resistant S. aureus (MRSA)
- Mupirocin inhibits bacterial isoleucyl transfer-RNA synthetase (IRS) and inhibits bacterial protein synthesis.
Resistance to mupirocin
- Low-level resistance is due to mutation of the host IRS, whereas high-level resistance is due to the acquisition of a distinct IRS that is less sensitive to inhibition
- The low-level mupirocin resistance involves point mutations in the ileS gene and High-level mupirocin resistance is mediated by the plasmid-encoded genes mupA and mupB.
References
- https://pubmed.ncbi.nlm.nih.gov/26165756/
- https://pubmed.ncbi.nlm.nih.gov/11807177/
- https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4752126/
- https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4313920/
- https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4817740/
- https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1114060/
- https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4626314/
- https://pubmed.ncbi.nlm.nih.gov/15013035/
- https://aac.asm.org/content/56/2/1001
- https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4515024/
- https://aac.asm.org/content/58/4/2281
- https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4158649/