Contents:
Introduction
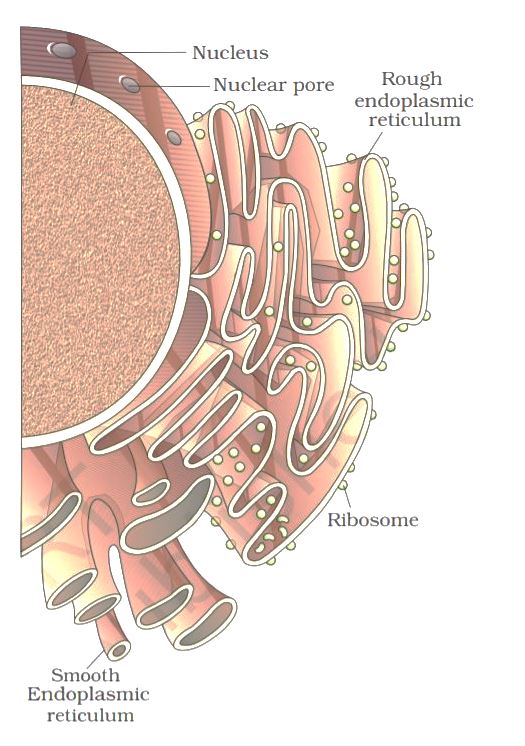
The endoplasmic reticulum (ER) is organized into a netlike labyrinth of branching tubules and flattened sacs that extend throughout the cytosol. The tubules and sacs interconnect, and their membrane is continuous with the outer nuclear membrane. The endoplasmic reticulum is entirely enclosed by a continuous membrane and is the largest organelle of most eukaryotic cells.
There are three contiguous membrane domains within the Endoplasmic reticulum that perform different functions within the cell. The rough ER is covered by ribosomes on its outer surface, and the transitional ER, where vesicles exit to the Golgi apparatus, both function in protein processing. The smoothER is not associated with ribosomes and is involved in lipid, rather than protein, metabolism.
The Endoplasmic Reticulum and Protein Secretion
The Endoplasmic Reticulum is the site known for protein secretion and protein sorting. the proteins after sorting move to their target region in a fixed pathway. The pathway could be explained as the secretory pathway: Rough ER → Golgi apparatus → secretory vesicles → cell exterior. The secretory proteins traveled from the Golgi apparatus to the cell surface in secretory vesicles, which then fused with the plasma membrane to release their contents outside of the cell.

Targeting Proteins to the Endoplasmic Reticulum
In higher eukaryotic cells, the sorting of proteins takes place simultaneously with the translation process. The free ribosomes synthesize the protein and transport it to the nucleus, mitochondria, peroxisomes, and chloroplasts while the membrane-bound ribosomes synthesize and transport it to the plasma membrane, secretory vesicles, lysosomes, and endosomes. They may be either retained within the ER or transported to the Golgi apparatus and, from there, to lysosomes, the plasma membrane, or the cell exterior via secretory vesicles.
Insertion of Proteins into the ER Membrane
Proteins destined for secretion from the cell or residence within the lumen of the ER, Golgi apparatus, endosomes, or lysosomes are translocated across the ER membrane and released into the lumen of the ER. However, proteins destined for the plasma membrane are inserted into the ER membrane. From the ER membrane, they proceed to their final destination along the same pathway as that of secretory proteins: ER → Golgi → plasma membrane or endosomes → lysosomes.
Integral membrane proteins span the membrane via α-helical regions of 20 to 25 hydrophobic amino acids, which can be inserted in a variety of orientations. However, different integral membrane proteins vary in how they are inserted. Some integral membrane proteins span the membrane only once, while others have multiple membrane-spanning regions
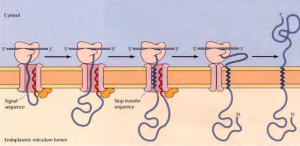
Insertion of a membrane protein with a cleavable signal sequence and a single stop-transfer sequence – The signal sequence is cleaved as the polypeptide chain crosses the membrane, so the amino terminus of the polypeptide chain is exposed in the ER lumen. However, translocation of the polypeptide chain across the membrane is halted by a transmembrane stop-transfer sequence that closes the translocon and exits the channel by anchoring the protein in the ER membrane.
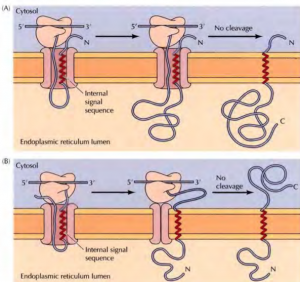
Insertion of membrane proteins with internal non-cleavable signal sequences- Internal non-cleavable signal sequences can lead to the insertion of polypeptide chains in either orientation in the ER membrane. The signal sequence directs the insertion of the polypeptide such that its amino terminus is exposed on the cytosolic side. The remainder of the polypeptide chain is translocated into the ERas translation proceeds. The signal sequence is not cleaved, so it acts as a membrane-spanning sequence that anchors the protein in the membrane with its carboxy terminus in the lumen of the ER.
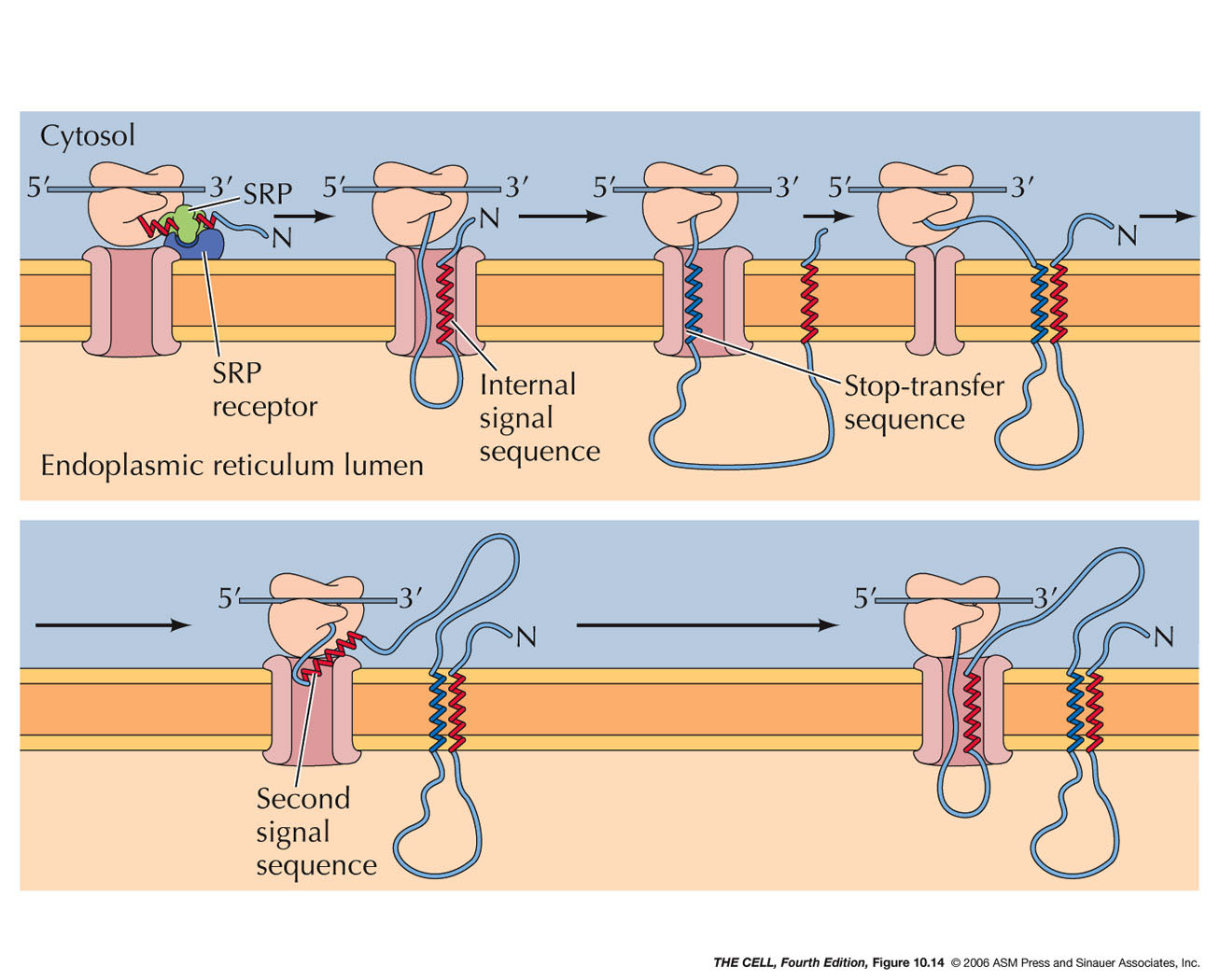
Insertion of a protein that spans the membrane multiple times- An internal signal sequence results in the insertion of the polypeptide chain with its amino terminus on the cytosolic side of the membrane. A stop-transfer sequence then signals the closure of the translocation channel causing the polypeptide chain to form a loop within the lumen of the ER; translation continues in the cytosol. A loop is formed in the cytosol when a second internal signal sequence reopens the channel, triggering the reinsertion of the polypeptide chain into the ER membrane. The process can be repeated many times, resulting in the insertion of proteins with multiple membrane-spanning regions.
Function Of Endoplasmic Reticulum
1. Protein Folding and Processing in the ER
ER involves the folding of polypeptide chains into their correct three-dimensional conformations, the assembly of polypeptides into multisubunit proteins, and the covalent modifications. Polypeptide folding is assisted by molecular chaperones Hsp70 and BiP which binds to the unfolded polypeptide chain as it crosses the membrane and then mediates protein folding and the assembly of multisubunit proteins within the ER. The correctly assembled proteins are released from BiP and are transported to the Golgi apparatus. Abnormally folded or improperly assembled proteins are targets for degradation.
The formation of disulfide bonds between the side chains of cysteine residues is supported within the ER due to the oxidizing environment. These bonds do not form in the cytosol, due to the reducing environment. Disulfide bond formation is facilitated by the enzyme protein disulfide isomerase which is located in the ER lumen. The BiP not only helps in protein folding but also binds to the signaling molecules to avoid interference in the protein folding process.
2. Protein Glycosylation
Proteins are also glycosylated on specific asparagine residues (N-linked glycosylation) within the ER while their translation is still in process.
3. Quality Control in the ER
The proteins that fail to fold correctly in the ER are degraded while; others reside in the ER for several hours as they are properly folded. Thus an important role of the ER is to identify misfolded proteins, mark them, and divert them to a degradation pathway. Because they assist proteins incorrect folding, chaperones, and protein processing enzymes in the ER lumen, they often act as sensors of misfolded proteins. The process of ER protein quality control is complex and involves at least four chaperones, protein disulfide isomerase, and many supporting proteins. In addition to acting as a chaperone, BiP plays a pivotal role as a sensor of the general state of protein folding within the cell.
Despite all the help from chaperones, many protein molecules (more than 80% for some proteins) translocated into the ER fail to achieve their properly folded or oligomeric state. Such proteins are exported from the ER back into the cytosol, where they are degraded. The mechanism of retrotranslocation, also called dislocation, is still unknown but is likely to be similar to other post-translational modes of translocation
The Smooth ER and Lipid Synthesis
The SER is the major site at which membrane lipids are synthesized in eukaryotic cells. Because they are extremely hydrophobic, membrane lipids are synthesized in association with already existing cellular membranes rather than in the aqueous environment of the cytosol. Although some lipids are synthesized in association with other membranes, most are synthesized in the ER. The proteins are then transported to their targeted regions either in vesicles or by carrier proteins. The membranes of eukaryotic cells are composed of three main types of lipids: phospholipids, glycolipids, and cholesterol.
Synthesis of Phospholipids
Most of the phosphoGlycerol phospholipids are synthesized in the ER membrane from cytosolic precursors. Two fatty acids Linked to coenzyme A (CoA) carriers are first joined to glycerol-3-phosphate, yielding phosphatidic acid, which is simultaneously inserted into the membrane. A phosphatase then converts phosphatidic acid to diacylglycerol. The attachment of different polar head groups to diacylglycerol then results in the formation of phosphatidylcholine, phosphatidylethanolamine, or phosphatidylserine. Phosphatidylinositol is formed from phosphatidic acid via a modified diacylglycerol
Translocation of phospholipids across the ER membrane
Because new lipid molecules are added only to the cytosolic half of the bilayer and lipid molecules do not flip spontaneously from one monolayer to the other, a membrane-bound phospholipid translocator (called a scramblase) is required to transfer lipid molecules from the cytosolic half to the lumenal half so that the membrane grows as a bilayer. The scramblase is not specific for particular phospholipid head groups and therefore equilibrates the different phospholipids between the two monolayers
Export of Proteins and Lipids from the ER
Proteins and lipids are carried from the ER to the Golgi in transport vesicles that bud from the membrane of the transitional ER, fuse to form the vesicles and tubules of the ER-Golgi intermediate compartment (ERGIC), and are then carried to the Golgi. Lumenal ER proteins are taken up by the vesicles and released into the lumen of the Golgi. Membrane proteins maintain the same orientation in the Golgi as in the ER
References
- Cell and Molecular Biology by Karp 5th Ed
- Karp’s Cell and Molecular Biology: Concepts and Experiments, 5th Edition Gerald Karp, Janet Iwasa, Wallace Marshall
- https://www.ncbi.nlm.nih.gov/books/NBK9889/
- https://www.ncbi.nlm.nih.gov/books/NBK26841/