Probes are short section of DNA or RNA with an additional tagged or labeled chemical entity that are used to bind to its complimentary strand and thereby allows detection of candidate nucleic acid molecule. A fluorescence molecule or a colour bead or quantum dots (Cd-Se Qdots, Zn SQdots), photochromic compounds, isotopic labelling or non-isotopic labelling, etc. can be used for this chemical-synthesized entity. It helps us to view when a sample binds to DNA , RNA and other target nucleic acids.
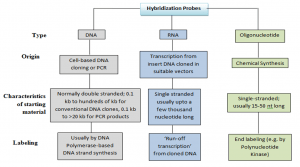
Labeling Of Probes:
- Samples may be labelled internally at different locations. or in a certain position in the oligonucleotides . Some probes are of defined length and some are heterogeneous populations of labelled molecules.
- In vivo labelling: In tissue cell culture cell DNA and RNA can be directly labelled in vivo by inserting deoxynucleotides. This method is restricted only to prepare labelled viral DNA from virus-infected cells and to study RNA processing events.
- In vitro labelling: This is a more flexible technique requiring the use of DNA polymerases for labelling in-vitro of purified RNA , DNA or oligonucleotide.
Labeling during synthesis:
In vitro labeling of DNA can be done by various methods as follows:
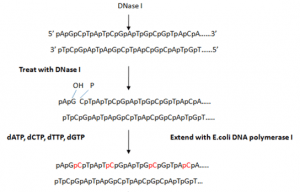
a) Nick-translation
- Nick translation is a radioactive process for the labelling of two-stranded DNA that makes it suitable for the detection of specific genomic sequences.
- It involves insertion of random single-strand breaks called nicks in one of the strands of double stranded target DNA which exposes 3′- OH termini and 5′-PO4 termini. The nicks are introduced by endonuclease like pancreatic deoxyribonuclease I (DNase I).
- Addition of DNase I and multi-subunit enzyme E. coli DNA polymerase I is used for nick translation which contributes both activities like:
(i) 5′ → 3′ exonuclease that attacks the exposed 5′ termini of a nick and sequentially removes the nucleotides in 5′ → 3′ direction
(ii) DNA polymerase adds nucleotides to the free 3′-OH group, in 5′ → 3′ direction replacing the nucleotides removed by exonuclease and causing lateral displacement (translation) of the nick. This method requires 100-fold less radioactive precursor than in-vivo labeling method. The amount of radiolabel incorporated depends on number of nicks created by DNase I.
- At low temperatures (approx. 15 ° C), it is a drawback that there is only one overall regeneration of the current nuclear chain and no further reaction.
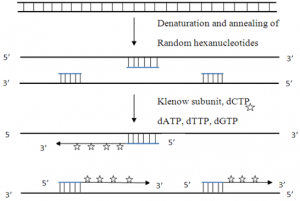
b) Random primed labeling
- The techniue uses short random sequence hexanucleotides that prime at several locations with denatured target DNA.
- This method known as oligo-labeling is based upon hybridization of a mixture of all possible hexanucleotides.
- The template DNA is initially denatured and then cooled slowly so as to allow random hexanucleotides to bind at complementary sequences at which extension takes place through PCR.
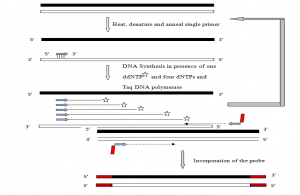
c) PCR-mediated labeling
- Labeling of RNA is generally accomplished by an in vitro transcription system. These procedures require DNA or RNA polymerases to add labeled nucleotides to synthesize in vitro probes. It requires at least one labeled nucleotides among four nucleotides.
This method has several advantages over other methods
- Identified target DNA segments may be amplified and labelled regardless of the restricting sites.
- DNA template requirement is too small
- The isolation is not required for DNA fragments or sub-cloning in bacteriophage promoters containing vectors
The standard PCR reaction can be modified to incorporate labeled nucleotides. The methods generally used are
- Standard PCR-based DNA labeling- The probe generation reaction is modified to incorporate one or more labeled nucleotide precursors at a concentration equivalent as oligonucleotide conc. (Km) or slightly above Km and others at concentrations surpassing Km, which become incorporated into the PCR product throughout its length.
- Primer-mediated 5′ end labeling- This method uses a 5′ end-labeled primer, which is incorporated during the PCR reaction. It is utilized in DNA sequencing, PCR-based mutagenesis, etc. Radiolabeled probes can be generated for both strands using equal concentrations of primers or biased heavily in favor of one strand of DNA using a higher concentration of one primer.
Preparation of Labeled Nucleotides
Nucleotides can be labeled by isotopic and non-isotopic methods.
Isotopic labeling:
Isotopes generally used for labeling nucleotides are 32P, 33P, 35S or 3H. They can be detected directly in solution or on X-ray film using autoradiography.
Properties of radioisotopes used for labeling DNA and RNA probes:
- Autoradiographical signal strength depends on radiation intensity from radiation isotopes and exposure duration.
- 32P emit high-energy β-particles with a high sensitivity to detection. Therefore it is commonly used in southern blot, hybridise, dot blot, and colony hybridization.
- But it is relatively unstable and the image is unambiguous because of its high energy β-particle emission when a fine resolution is necessary to interpret results.
- This makes it possible to use 35S-labelled, 33P (moderate semiconductions) or 3H-labeled nucleotides that emit less energy. They are used for in situ hybridization and in DNA sequence. 3H needs a long period of exposure due to the release of β-particulate energy.
Non-isotopic labeling:
Non-isotopic labeling systems involve the use of nonradioactive probes. These methods are developed recently as compared to radioisotope labeling methods, but are finding wide variety of applications in different ways. Two types of non-radioactive labeling are conducted: direct and indirect.
- Direct non-isotopic labeling – Here a nucleotide containing label such as Fluorescein, Texas Red, Rhodamine will be detected when incorporated with the help of spacer molecule. These modified nucleotides having tag, fluoresce when excited by light of certain wavelength.
- Indirect non-isotopic labeling- This involves a chemical link of reporter molecule to a nucleotide. When this modified nucleotide is incorporated into DNA, then it is specifically bound to a protein or other ligand which has high affinity against the reporter group. Long spacer is introduced between nucleotide and reporter so as to reduce steric hindrances for binding of affinity molecule.
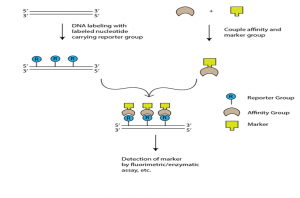
Two commonly used types of non-isotopic labeling are:
- Biotin-Streptavidin Method : This method uses two ligands with a high affinity: biotin acts as a reporter and streptavidine as the molecule of affinity is the bacterial protein. The labelling agents used are biotinylated nucleotites like bio-11-dUTP with a spacer of 4- 16C long atoms between biotin and dNTP . Biotin is an omnipresent part of mammalian tissues and is also added easily to some nylon membranes, which contributes to high background levels during hybridisation at place, northern and southern level. Digoxigenin is used to solve this challenge.
- Digoxigenin- A plant steroid is used as a reporter and molecule of affinity. Digoxigenin therefore is an all-purpose immuno tag and, in particular, a typical in situ hybridization histochemical marker.
Detection of non-radioactively labeled probes after hybridization
- Associated with a variety of marker groups or molecules are affinity molecules (streptavidin, digoxigenin-specific Antibodies). These contain a range of flowers or enzymes, including alkaline phosphatase and peroxidase, which can be identified by colorimetric testing, chemical luminescence testing and fluorescent testing.
- The colorimetric experiments are used to produce a phosphate compound dimerizing into the di-bromo-di-chloro-indigo, which reduces the NBT (nitrobluetetrazolium) to insoluble purple colour, which is noticeable on locations at which the probe has been hybrid.
- Fluorescent assays- HNPP (2-hydroxy-3-naphthoic acid 2′-phenylanilide phosphate) was used in fluorescent assays. HNPP produces fluorescent precipitate on membranes, which can be excited by irradiation at 290 nm, after de-phosphorylation of alkaline phosphatase. CCD cameras record the response signal provided at 509 nm.
- Chemiluminescence- The easiest and most sensitive test is chemiluminescence using the luminal detection system HRP (Horseradish Peroxidase). HRP catalyses the luminal oxidation in the presence of H2 O2, forming reactive peroxide that emits light in its ground state at 425 nm during decomposition.
References
- https://geneticeducation.co.in/dna-probes-labelling-types-and-uses/
- https://www.sciencedirect.com/science/article/pii/S0075753508700780
- https://nptel.ac.in/content/storage2/courses/102103013/pdf/mod3.pdf
- https://cdn-cms.f-static.net/uploads/3178028/normal_5e758be52b3a5.pdf
- https://www.scribd.com/document/369959460/Fundamentals-and-Techniques-of-Biophysics-and-Molecular-Biology