Contents:
pET and pBAD expression systems
EXPRESSION SYSTEMS
Fortunately, not only plasmid vectors containing promoter and ribosome binding sites located just before one or more restriction sites that allow easy insertion of the DNA encoding of the protein of interest have been developed, they have also been made commercially available. Indeed, a wide range of expression systems based on different promoters are available. Table 1 shows some of the commonly used systems, including the T7-based pET expression system (marketed by Novagen) and one based on the araBAD promoter (e.g. Invitrogen pBAD), which are discussed below.
The Expression System of PET
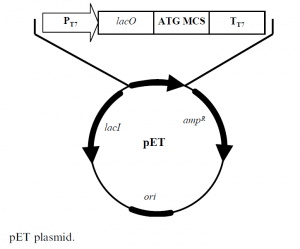
This system has been developed for a variety of expression applications, including hybrid promoters, multiple cloning sites for the incorporation of different fusion partners and protease cleavage sites. The pET plasmid, shown in Figure 1, is a 5.4 kb construct with the following elements:
- lacI codes for the lac repressor protein
- ampR codes for ampicillin resistance
- ori col E1 origin of replication
- lacO codes for the lac operon
- PT7 codes for the T7 promoter, which is specific only for T7 RNA polymerase
- TT7 codes for the T7 terminator
- MCS multiple cloning site, has the sequence encoding the protein of interest.
The use of PT7, a 20-nucleotide sequence that the E does not recognise, is a key feature. In the prokaryotic genome sequence, coli RNA polymerase does not occur, nor does T7 RNA polymerase; therefore, PT7 is not activated in the absence of T7 RNA polymerase and no protein of interest is produced. As described below, when PT7 is activated as a viral promoter, it transcribes quickly, at a maximum speed of around 230 nucleotides per second, some five times faster than E. Coli Polymerase RNA. Expression requires a host strain under the control of the IPTG inducible lacUV5 promoter lysogenized by a DE3 phage fragment, encoding the T7 RNA polymerase, and a schematic of the process is shown in Figure 2.
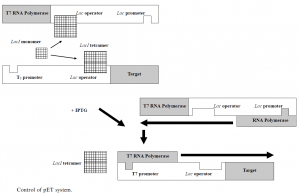
There is a copy of the lacI gene found on the E. Genome of coli, and pET. LacI is a weakly expressed gene and a 10-fold enhancement of the repression is achieved when the overexpressing promoter mutant LacIq is employed.35 The lac repressor protein, LacI, represses the lacUV5 promoter of the host cell and the T7 / lac hybrid promoter encoded by the expression plasmid. The lacI tetramer binds to the lac operator on both the host cell genome and the plasmid in the absence of an inducer then. This prevents the host cell from producing RNA polymerase T7 and prevents the protein of interest from being produced by the plasmid. It binds and causes the release of tetrameric lacI from the lac operator on both the genome and the plasmid when the inducer, typically IPTG, is introduced, which triggers the expression of T7 RNA polymerase in the host cell. In this way, transcription of the target gene from the hybrid promoter T7 / lac is initiated.
Low background expression from pET expression plasmids may occur; this may be reduced by co-expression with either plasmid pLysS or pLysE of T7 lysozyme, a natural T7 RNA polymerase inhibitor. With regard to the cognate tetracycline responsive (Tc) promoter, these plasmids harbour the T7 lysozyme gene in silent (pLysS) and expressed (pLysE) orientations.
Although the lacUV5 promoter is less sensitive than the lac promoter to the regulation of the cAMP-CRP (cAMP receptor protein) complex, the incorporation of 1% glucose into the culture medium reduces cAMP levels and significantly improves the promoter’s repression. Control of target protein expression is improved by host strains deficient in the lacY gene , which encodes lactose permease.
The pBAD Expression System
The regulation of the arabinose operon in E.coli is directed by the arabinose gene product38, which controls the rate of synthesis of the proteins AraE, AraF and AraG required for the absorption of arabinose, as well as the enzymes AraB, AraA and AraD required for its catabolism. That implies that the ara-specific promoters’ intrinsic state is off and AraC turns them on, while the lac operon promoter’s set state is on and the lac repressor turns it off. Moreover, pBAD is repressed by catabolites, This implies that growing the culture will further repress expression in the presence of glucose. While expression of a cloned plasmid gene containing the araBAD promoter can be modulated in cultures grown in the presence of sub-saturating arabinose concentrations over several orders of magnitude, individual cells are either fully induced or uninduced.
Cells having the arabinose transport gene (araE) that is natively controlled are either induced or uninduced, the relative fraction of which is controlled by arabinose concentration. As a function of inducer concentration, the population-averaged variation in pBAD expression is proportional to the percentage of cells that are fully induced (versus uninduced) rather than the level of expression in individual cells. In E.coli, this all-ornone phenomenon, which may have undesirable effects on heterologous gene expression, can be eliminated by araE expression from arabinose-independent promoters.
All cells in the population have approximately the same induction level in these arabinose-transport engineered cells.40 Therefore, strains capable of transporting L-arabinose but not metabolising it, such as a strain of recA, endA, are most appropriate.
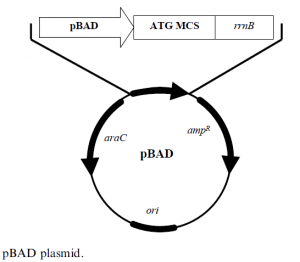
The araBAD promoter based expression plasmids are designed for tight background expression control and precise control of the target protein’s expression levels. This contrasts with the all-ornothing induction that most other systems of bacterial expression have achieved. A plasmid pBAD, derived from pBR322 and shown in Figure 3, is a construct of 4.1 kb that has the following elements:
- araC ORF encoding araC protein
- ampR codes for ampicillin resistance
- ori col E1 origin of replication
- pBAD araBAD promoter
- rrnB transcription termination region
- MCS multiple cloning site, has the sequence encoding the protein of interest.
In the arabinose operon, the AraC dimer binds three sites, I1, I2 and O2. In the absence of arabinose, 210 base pairs upstream from pBAD, the AraC dimer contacts the O2 site located within the araC gene. As shown, the other half of the araC dimer contacts the I1 site in the promotor region that forms the DNA loop. Therefore, transcription from pBAD and the promoter of araC (pC) is inhibited by the loop. The araC dimer changes its conformation upon binding of arabinose so that it binds to the pBAD I2 site instead of the O2 site. This removes the loop structure and transcription by RNA polymerase launches. The cAMP receptor protein (CRP) stimulates the binding of the araC dimer to the I1 and I2 sites, which means that glucose-mediated catabolite repression can reduce the background expression of araBAD.
REFERENCES:
- https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7205610/
- https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4029002/