Structure of RNA and its types
- Ribonucleic acid (RNA) is structurally similar to deoxyribonucleic acid (DNA), except for two major differences.
- It has sugar ribose instead of deoxyribose and nitrogenous base uracil in place of thymine in DNA. RNA is the polymer of four nucleotides each one contains D-ribose, phosphoric acid and a nitrogenous base.
- The bases are two purines (A and G) and two pyrimidines (C and U). RNA is usually a single-stranded polynucleotide. However, this strand may fold at certain places to give a double-stranded structure.
- There is no specific relation between purine and pyrimidine as well as guanine and cytosine contents due to the single-stranded nature of RNA.
- Ribonucleic acid (RNA) is present as their genetic material in some plant viruses (e.g. TMV, wound tumour viruses etc.), animal viruses (e.g. influenza viruses, foot and mouth viruses, poliomyelitis viruses etc.), and bacteriophages (e.g. MSz etc.).
- The organisms which have only RNA and it involved in genetic mechanism is called genetic RNA. The genetic RNA of viruses is self-replicating, that is, it can produce by its own replication.
- This mode of replication is called RNA-dependent RNA synthesis. DNA acts as a genetic material and RNA follows the order of DNA. In such cells the RNA does not have genetic role, is called non-genetic RNA is not self-replicating like DNA but it depends on DNA.
- Therefore, replication of non-genetic RNA is known as DNA-dependent RNA replication. Non-genetic RNA can be divided into three types on the basis of structure and function such as messenger RNA (mRNA ribosomal RNA (FRNA) and transfer RNA (tRNA).
(1) Messenger RNA:
- Francis Jacob and Jacques Monod (1961) proposed the name informational or messenger RNA (MRNA) for bearing the transcripts of DNA for protein synthesis on ribosomes.
- The molecule of mRNA is single-stranded like the rRNA molecule. Its base composition is similar like DNA so that GC contents of mRNA correspond to the GC contents of the genomes total DNA.
- The total population of mRNA in a cell varies from 5 to 10% of the total cellular RNA. The species of mRNA are short-lived and they are broken into ribonucleotides by the enzyme ribonuclease.
- The sedimentation coefficient of mRNA is 85 and its molecular weight ranges from 5,00,000 to 1,00,000. The mRNA found in prokaryotes shows different properties from that of eukaryotes.
- In prokaryotic, mRNA is complementary to chromosomal DNA and it forms RNA-DNA hybrids after the separation of the two DNA strands.
- Synthesis of mRNA is accomplished with one strand of DNA and that is used as a template. The enzyme RNA polymerase joins ribonucleotides and catalyzes the formation of 3′-5′-phosphodiester bonds that form the RNA backbone.
- The synthesis of mRNA is initiated at the 5′ end and it grows from 5′ end to 3′ end. The RNA polymerase attaches to an initiator site of the structural gene, in the promoter and it catalyzes mRNA synthesis.
- In eukaryotic, the synthesis of mRNA begins with the production of long precursor molecules by RNA polymerase II from the template strand of DNA.
- This enzyme function catalyzing the formation of 5’→3′ phosphodiester bonds of the RNA backbone by reading the DNA template in the 1’→5′ direction The developing MRHA is antiparallel and its nucleotides are complementary to those of the DNA template strand.
- The eukaryotic mRNA is capped at 5′ terminal end by 7 methyl guanosine triphosphate. This cap helps to prevent hydrolysis of miRNA by 5 exonucleases and it is also involved in the recognition of mRNA protein synthesis.
- The 3′ terminal end of miRNA contains a polymer of adenylate residues (onto 250 nucleotides) which is known as poly(A) tail and it provides stability to mRNA.
(2) Ribosomal RNA (rRNA):
- Ribosomal, stable or insoluble RNA constitutes the largest part (80%) of the total cellular RNA. In prokaryotes RNA is formed on a part of DNA called ribosomal DNA, while in eukaryotes it is formed in nucleolus containing the nuclear DNA.
- The RNAs are found in ribosomes. The ribosomes are of different types such as the 80S(eukaryotes) and 70S (prokaryotes). The 80S ribosome consists of 60S and 40S subunits. The 60S ribosomal subunit contains 28S rRNA, 5.8S tRNA, and 5S rRNA while the 40S subunits contain 18S rRNA, The 70S ribosomes consist of 50S and 30S subunits. The 50S subunit contains 23S and 5S rRNA while the 30S subunit consists of 16S rRNA.
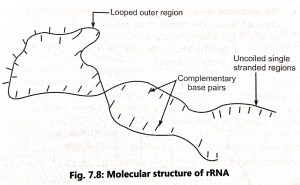
- The rRNA is a single stranded molecule which is twisted at some points to form helica regions. Most of the base pairs are complementary and linked by hydrogen bonds in the helical region (Fig. 1).
- The uncoiled single stranded regions lack the complementary bases Hence, the ratio of purine: pyrimidine is not equal. The rRNA may play a significant role the binding of mRNA to ribosomes and protein synthesis.
(3) Transfer RNA (tRNA):
- Transfer RNA or soluble RNA molecule contains 71 to 80 nucleotides with molecular weight ranging from 25,000 to 30,000 D.
- It is smaller RNA than mRNA and rRNA and has a sedimentation coefficient of 4S. It accounts for 10 to 20% of the total cytoplasmic RNA.
- In addition to A, G, C and U bases, some unusual bases are found in tRNA. The unusual bases are 3-methyl cytosine, inosine, dihydrouracil, methyl guanine, dimethyl guanine, ribothymine, pseudouridine, methylinosine etc.
- These unusual bases are mainly formed by chemical modification by common bases. Transfer RNA molecule actively engaged in protein synthesis is typically ‘clover leaf’ like in shape due to intrachain pairing of complementary nucleotides in certain region of the tRNA chain (Fig. 2).
- Robert Holley (1968) prepared the cloverleaf model for yeast tRNA alanine which includes several known functions of tRNA. A typical structure of tRNA shows the following features:
1. The single polynucleotide chain of all the tRNA molecules is folded upon itself to form five arms. e.g. acceptor or end terminus arm, dihydrouridine or DHU or D arm, anticodon arm, variable arm, and TψC arm.
2. All molecules have guanine residue ‘G’ at the 5′ terminal end and unpaired (single-stranded) – CCA sequence at the 3′ end. This is called amino acid attachment site or acceptor’s arm. Adenylic acid (A) acts as an amino acid attachment site because amino acids are covalently attached to the CCA sequence during polypeptide synthesis.
3. The acceptor arm consists of 7 base pairs and 4 unpaired bases. The unpaired bases contain a three – CCA bases and a forth variable purine (A or G) at 3′ end.
4. The dihydrouridine (DHU) or D loop constitutes 8 to 12 unpaired bases. It consists of 15 to 18 nucleotides (3 to 4 base pairs and 8 to 12 unpaired bases) in the loop. It acts as the site for recognition of amino acid activating enzyme aminoacyl tRNA synthetase.
5. The anticodon sterm contains five paired bases and anitcodon loop consists of seven unpaired bases. The middle three nucleotides (from unpaired bases, 3, 4 and 5 act as anticodon which identify three complementary bases of mRNA molecule. This interaction decides about the order of amino acids on polypeptide chain during assemblage in the ribosomes.
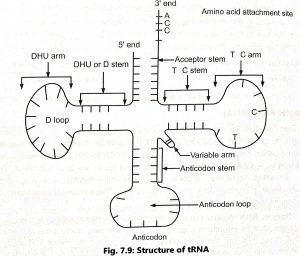
6. A variable arm or extra loop may be present in some tRNA which varies in shape, nucleotide content, and composition from tRNA to tRNA. It is mainly present in between the anticodon arm and T ψC arm.
7. TψC arm consists of a stem of 5 base pairs and a loop of 7 unpaired bases. The TψC loop consists of a TYC sequence at 5′ 3′ direction, It is involved in the binding of tRNA molecules to the ribosomes.
8. The tRNA that initiates protein synthesis is called initiator tRNA. The tRNA specifies methionine as the starting amino acid in eukaryotic protein synthesis and N-methyl methionine in prokaryotes. Hence, are methionyl RNA.
9. Three-dimensional structure (TDS) was proposed by S.H. Kim (1973) for yeast phenylalanine tRNA molecule. This TDS takes the shape of the letter ‘L’ with a thickness of two tRNAs specific to these two amino acids of 20A°.
REFERENCES
