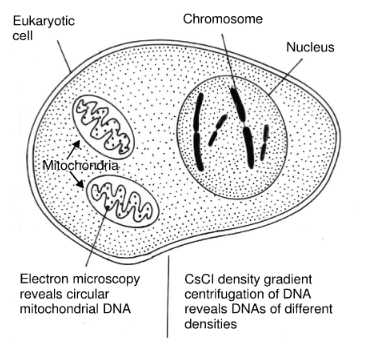
Mitochondria
Mitochondria are found in the cytoplasm of all animal and plant cells. are the principal source of energy and are also called as powerhouse of cells. The presence of DNA in mitochondria has been shown by a variety of techniques, including electron microscopy, autoradiography. They contain the enzyme of the Kreb’s cycle, carry out oxidative phosphorylation, and are involved in fatty acid biosynthesis.
Structure
- Mitochondrial DNA is a double-stranded, supercoiled, circular chromosome devoid of histones and therefore not organized into nucleosomes.
- The size of the chromosome varies from organism to organism but is constant among mitochondria within any particular species.
- Interestingly, the chromosomes of higher animal mitochondria are significantly smaller than those of fungi and plants.
- The mitochondrial chromosome in humans, for example, is about 14000 base pairs, while that from Drosophila is 18000 base pairs, that from Neurospora crassa is 60000 base pairs. and that from yeast is 75000 base pairs.
- The mitochondrial genomes of plants are even larger ranging from 250000 to 2 million base pairs (80 to 800 pm). depending on the species. Maize, for example. has mitochondrial genome of 600000 base pairs (200 pm).
- Mitochondrial genomes are much more variable in terms of size and topology. The genome sizes of known mitochondrial DNAs vary over a range of several orders of magnitude from ten million to over a billion relative molecular weight.
- Since the information encoded by the mitochondrial genome appears to be very much the same in all organisms, the size heterogeneity must reflect either the presence of non-coding DNA. or the presence of multiple copies of coding sequence.
Replication
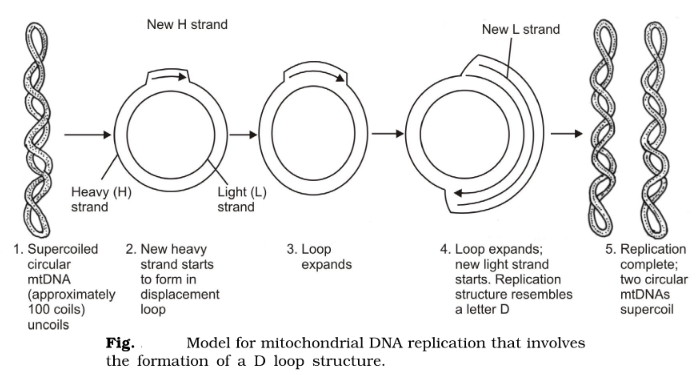
Replication of mtDNA used mitochondrial DNA polymerases that function totally independently of nuclear DNA polymerases. Moreover, the replications process occurs throughout the cell cycle without any preference for the S phase of the cell cycle when nuclear DNA replicates. Some differences appear in the details of mtDNA replication among eukaryotes, and the displacement loop (D loop) model (deduced from observing animal mitochondria in vivo) is useful as a general scheme.
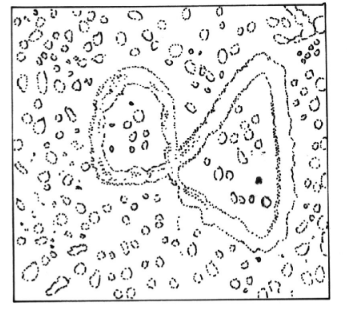
In most animals, the two strands of mtDNA have different densities, and they are called the H (heavy) and L (light) strands. The D loop model of replication shows relative asynchrony in DNA replication for the two complementary H and L strands. In the D loop model, the synthesis of the new H strand is started in one origin (the L strand origin) and forms a D loop structure (which can be seen by electron microscopy). As the new H strand extends, initiation of synthesis of the new L strand at a second origin (the H strand origin) takes place. Both strands are completed by continuous replication. Lastly, the circular DNAs of each are converted to a supercoiled form with approximately a hundred superhelical twists.
As in the replication of nuclear DNA, RNA primers are synthesized for initiation. Apparently, the excision of these primers is not precise since interspersed ribonucleotides are found in the mtDNA. The evidence is that mtDNA can be cleaved by treatment with alkali or RNase, both of which are known to cleave at phosphodiester linkages in which a ribose sugar is involved. About ten of these ribonucleotides are found in each mtDNA, with most of them clustered around the replication origins.
Gene Organization of mtDNA
Mitochondrial DNA contains information for a number of mitochondrial components such as tRNAs, rRNAs, and proteins. Mitochondria contain ribosomes that are responsible for all protein synthesis activity that occurs within mitochondria. Messenger RNAs synthesized within the mitochondria remain in the organelle and are translated by mitochondrial ribosomes.
The two rRNA molecules found in mitochondria ribosomes are coded for in the mtDNA. while most, if not all, of the ribosomal proteins of mitochondrial ribosomes, are coded for in the nuclear genome. In general, mtDNA codes of 2 rRNAs, 22-23 (RNAs, and 10-12 proteins that are part of the inner mitochondrial membrane and are hydrophobic to some degrees. The mtDNA molecule has been studied in a variety of organisms, and since it is relatively small, a great deal of progress has been made in determining the exact organization of its genes.
One approach to mapping mtDNA physically has been through denaturation. The DNA is placed in a medium that causes stretches of AT base pairs to denature while GC-rich regions remain hydrogen-bonded. This selective denaturation leads to the formation of denaturation bubbles that can be visualized by electron microscopy. This physical map shows AT-rich regions of the DNA that can be used as landmarks in other studies.
The structural genes are also found in both strands. Their positions were identified in two ways. One method involved searching the DNA sequence for possible start and slop signals in the same reading frame (and open reading frame or OFR), they were separated by significant lengths and that could code for amino acids. This search was accomplished by using computer programs designed for this purpose.
The second method (used primarily by G. Attardi and his group) was to align the 5’ and 3′ end proximal sequences of mitochondrial mRNAs with the DNA sequence. The mitochondrial mRNAs were isolated and were found to have a poly (A) tail at the 3’ end but no 5′ cap. As in the case of cytoplasmic mRNAs (i.e., the mRNAs coded for by nuclear genes but found in the cytoplasm), the poly (A) tail is a great aid in isolating these molecules.
