Types of Blotting Techniques
Blotting techniques are widely used analytical tools for the specific identification of desired DNA or RNA fragments. Blotting refers to the process of immobilization of sample nucleic acids or solid support. The blotted nucleic acids are then used as targets in the hybridization experiments for their specific detection.
Southern blotting:
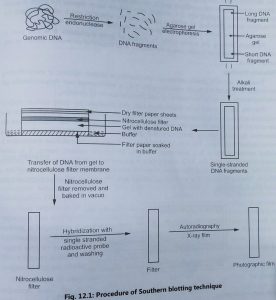
- The original method of blotting was developed by E. M. Southern (1975) to identify the locations of genes and other DNA sequences on restriction fragments separated by gel electrophoresis.
- In Southern blotting technique, a sample of DNA containing fragments of different sizes is subjected to electrophoresis using either polyacrylamide or agarose gel.
- DNA molecule is cut into small fragments by restriction enzyme and passed through agarose gel by electrophoresis method. It results in the separation of DNA molecules based on their size.
- The DNA is then denatured into single strands by exposing the gel to an alkaline solution. The gel is added on the top of the buffer saturated filter paper.
- It is covered with a nitrocellulose filter overlayed with dry filter paper (Fig. 1). The buffer moves from the bottom of filter paper by the capillary action and DNA is trapped in the nitrocellulose membrane.
- This process is known as blotting. The nitrocellulose membrane is removed from the blotting stack and DNA is permanently immobilized on the membrane by baking at 80°C or ultraviolet-induced cross-linking.
- Single-stranded DNA has a high affinity for nitrocellular filter membranes. Hence, the membrane is treated with a solution containing 0.2% each of Ficoll, polyvinylpyrrolidone and bovine serum albumin.
- This treatment prevents non-specific binding of the radioactive probe. The membrane is placed in a solution of labelled RNA, single-stranded DNA or oligodeoxynucleotide (probe).
- This labelled nucleic acid is used to detect and locate the complementary sequence, it is called a probe.
- The probe containing the sequence of interest is hybridized or annealed with the immobilized DNA on the membrane.
- After the hybridization reaction, the membrane is washed to remove the unbound probes. The washed membrane is exposed to X-ray film that detects the presence of the radioactivity in the bound probe.
- The film is developed to reveal bands indicating positions in the gel of the DNA fragments that are complementary to the radioactive probe.
- The Southern blotting technique is very sensitive method and it is used to map the restriction sites around a single copy gene sequence in any genome.
- It is also used preparation of restriction fragment length polymorphism (RFLP) maps, DNA finger printing identification of the transferred genes etc. Nitrocellulose paper being very fragile is non replaced by nylon membrane.
Northern blotting:
- Alwine et al (1979) devised a technique in which RNA bands are blot transferred from the gel onto chemically reactive paper. An aminobenzyloxymethyl cellulose paper was prepared from Whatman paper 540 after a series of simple reactions.
- It is diazotised and rendered into the reactive paper. It becomes available for hybridization with radio-labelled DNA probes.
- The hybridized bands are found out by autoradiography. Alwine’s method extends that of Southern method and hence, it has given the jargon term ‘Northern blotting’.
- Thomas (1980) found that mRNA bands can also be blotted directly on nitrocellulose paper under appropriate condition and it can be hybridized with a labelled DNA or RNA probe.
- Hybrids are treated with S-1 nuclease with RNAase which digests the single-stranded RNA/DNA probe.
- Structure of mRNA is revealed to the extent to which mRNA protects the nucleic acid probe, In this technique, preparation of reactive paper is not required.
- Northern blotting is mainly useful in studies of gene expression. It is also used to determine whether a particular gene is transcribed in all tissues of a microorganism or only certain tissues.
Western blotting:
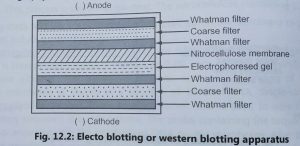
- Towbin et al (1979) developed the western blotting or protein blotting or electroblotting technique to find out the newly encoded protein by a transformed cell.
- Polyacrylamide gel electrophoresis is used for the separation and characterization of proteins.
- Proteins are extracted from transformed cells and separated by using sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS – PAGE).
- Sodium dodecyl sulphate acts as a denaturant for proteins during electrophoresis. After electrophoresis, individual proteins are detected by using specific antibodies and polypeptides can also be transferred to a nitrocellulose membrane.
- The transfer of proteins from gels to nitrocellulose membranes is called western blotting. It is performed by using an electric current, hence, it is called electroblotting method (Fig. 2).
- The electric field is applied to cause the migration of proteins from the gel to nitrocellulose filter paper. Nitrocellulose membrane is used for probing with a specific labelled antibody. The antibody is labelled with t and the signal is detected again with autoradiography.
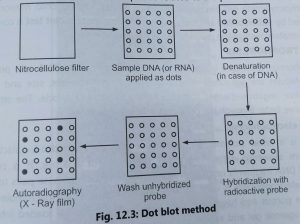
Dot and slot blotting:
- Dot and slot blotting is used to detect the presence of a given sequence of DNA/RNA In the non-fractionated DNA.
- The procedure is made simple by excluding purification steps electrophoresis and blotting of gel.
- The sample of DNA or RNA from different individuals or tissues are transferred onto a nitrocellulose filter in form of dots or slots, Hence, this method is called dot and slot blotting.
- The DNA is first denatured and then the filer is baked al 80°C to fix the DNA. The nitrocellulose filter is treated with the appropriate radioactive single-stranded DNA probe.
- The membrane is then hybridized with radioactively labelled probe. (DNA or RNA) and dots are detected by autoradiography (Fig. 3).
- The intensity of dot will indicate the relative concentration of a sequence related to the probe in the DNA sample.