Deoxyribonucleic acid (DNA)
It is a molecule composed of two chains that coil around each other to form a double helix carrying genetic instructions for the development, functioning, growth, and reproduction of all known organisms and many viruses.
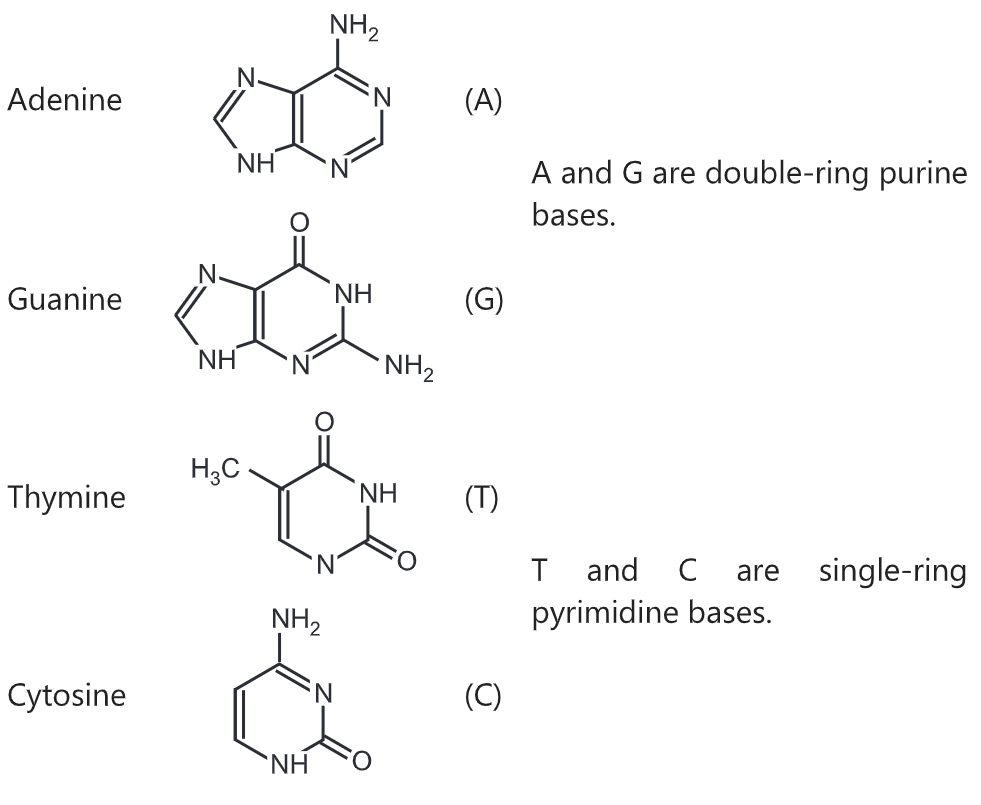
The two DNA strands are also known as polynucleotides as they are composed of simpler monomeric units called nucleotides. Each nucleotide is composed of one of four nitrogen-containing nucleobases (cytosine [C], guanine [G], adenine [A] or thymine [T]), a sugar called deoxyribose, and a phosphate group. The nucleotides are joined to one another in a chain by covalent bonds between the sugar of one nucleotide and the phosphate of the next, resulting in an alternating sugar-phosphate backbone. The nitrogenous bases of the two separate polynucleotide strands are bound together, according to base-pairing rules (A with T and C with G), with hydrogen bonds to make double-stranded DNA. The complementary nitrogenous bases are divided into two groups, pyrimidines, and purines. In DNA, the pyrimidines are thymine and cytosine; the purines are adenine and guanine.
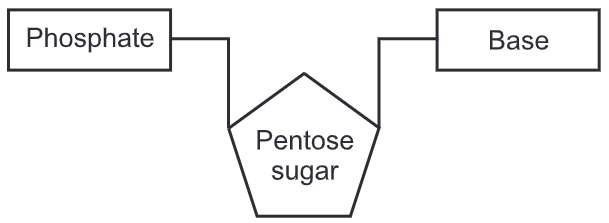
There are four organic bases involved in the formation of DNA molecules:
- adenine and guanine (both purines containing two rings in their structures)
- thymine and cytosine (both pyrimidines containing only one ring in their structures)
Both strands of double-stranded DNA store the same biological information. This information is replicated as and when the two strands separate. A large part of DNA (more than 98% for humans) is non-coding, meaning that these sections do not serve as patterns for protein sequences. The two strands of DNA run in opposite directions to each other and are thus antiparallel. Attached to each sugar is one of four types of nucleobases. It is the sequence of these four nucleobases along the backbone that encodes genetic information.
DNA is a long polymer made from repeating units called nucleotides. In all species, it is composed of two helical chains, bound to each other by hydrogen bonds. Both chains are coiled around the same axis, and have the same pitch of 34 angstroms (A) (3.4 nanometres). The pair of chains has a radius of 10 angstroms (1.0 nanometre).
DNA does not usually exist as a single strand, but instead as a pair of strands that are held tightly together. These two long strands coil around each other, in the shape of a double helix. The nucleotide contains both a segment of the backbone of the molecule (which holds the chain together) and a nucleobase (which interacts with the other DNA strand in the helix). A nucleobase linked to a sugar is called a nucleoside, and a base linked to a sugar and to one or more phosphate groups is called a nucleotide. A biopolymer comprising multiple linked nucleotides (as in DNA) is called a polynucleotide.
The backbone of the DNA strand is made from alternating phosphate and sugar residues. The sugar in DNA is 2-deoxyribose, which is a pentose (five-carbon) sugar. The sugars are joined together by phosphate groups that form phosphodiester bonds between the third and fifth carbon atoms of adjacent sugar rings. These are known as the 3′-end (three prime end), and 5′-end (five prime end) carbons, the prime symbol is used to distinguish these carbon atoms from those of the base to which the deoxyribose forms a glycosidic bond.
Any DNA strand therefore normally has one end at which there is a phosphoryl attached to the 5’ carbon of a ribose (the 5’ phosphoryl) and another end at which there is a free hydroxyl attached to the 3′ carbon of a ribose (the 3’ hydroxyl). The orientation of the 3’ and 5’ carbons along the sugar-phosphate backbone confers polarity to each DNA strand. In a nucleic acid double helix, the direction of the nucleotides in one strand is opposite to their direction in the other strand: the strands are antiparallel. The asymmetric ends of DNA strands are said to have a polarity of five prime end (5’).
The DNA double helix is stabilized primarily by two forces: hydrogen bonds between nucleotides and base-stacking interactions among aromatic nucleobases. The four bases found in DNA are adenine (A), cytosine (C), guanine (G), and thymine (1). These four bases are attached to the sugar-phosphate to form the complete nucleotide, as shown for adenosine monophosphate. Adenine pairs with thymine and guanine pairs with cytosine, forming A-T and G-C base pairs.
The nucleobases are classified into two types: the purines, A and G, which are fused five- and six-membered heterocyclic compounds, and the pyrimidines, the six-membered rings C and T.
There are four areas in which the structural forms of DNA can differ.
1. Handedness – right or left
2. Length of the helix turn
3. Number of base pairs per turn
4. Difference in size between the major and minor grooves
The tertiary arrangement of DNA’s double helix in space includes B-DNA, A-DNA, and Z-DNA.
(a) B-DNA is the most common form of DNA in-vivo and is a narrower, elongated helix than A-DNA. Its wide major groove makes it more accessible to proteins. On the other hand, it has a narrow minor groove. B-DNAs favoured conformations occur at high water concentrations and the hydration of the minor groove appears to favour B-DNA. B-DNA base pairs are nearly perpendicular to the helix axis.
(b) A-DNA is shorter and wider than helix B. Most RNA and RNA-DNA duplex are in this form. A-DNA has a deep, narrow major groove which does not make it easily accessible to proteins. On the other hand, its wide, shallow minor groove makes it accessible to proteins but with lower information content than the major groove. Its favored conformation is at low water concentrations. A-DNAs base pairs tilt to the helix axis and are displaced from the axis.
(c) Z-DNA is a relatively rare left-handed double-helix. Given the proper sequence and superhelical tension, it can be formed in-vivo but its function is unclear. It has a narrower, more elongated helix than A or B. Z-DNA’s major groove is not really groove and it has a narrow minor groove. The most favoured conformation occurs when there are high salt concentrations. There are some base substitutions but requires an alternating purine-pyrimidine sequence.
