Nucleic Acid Analysis Methods
There are numerous methods for DNA and RNA analysis, but many of them are solution-based or include the use of chip-based array systems more recently. Indeed, the laboratory-on-a-chip approach is developing rapidly and many detection and analysis methods can be envisaged to be developed in this format in the future. In many laboratories, however, traditional methods are still used and much is still done to produce a hard copy of digested and separated single stranded DNA fragments attached to a matrix such as nylon for analysis with a suitable labelled probe.
DNA Blotting
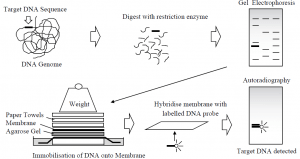
- DNA restriction fragment electrophoresis allows for size-based separation, but does not provide any indication as to the presence of a specific, desired fragment in the complex sample.
- By transferring the DNA from the intact gel to a piece of nitrocellulose or nylon membrane placed in contact with it, this can be achieved. Since DNA begins to diffuse from a gel that is left for a few hours, this provides a more permanent record of the sample.
- First, to render the DNA single stranded, the gel is soaked in alkali. It is then transferred to the membrane so that, in exactly the same pattern as that originally on the gel, the DNA is bound to it.
- This transfer can be performed electrophoretically or by drawing large volumes of buffer through both gel and membrane, named a Southern blot after its inventor Ed Southern, thus transferring DNA by capillary action from one to the other.
- The point of this operation is that a labelled DNA molecule, such as a gene probe, can now be used to treat the membrane. Under the right conditions, this single-stranded DNA probe will hybridise into complementary fragments immobilised on the membrane.
- For this process to take place effectively, the conditions of hybridisation, including the temperature and salt concentration, are crucial. This is usually referred to as the hybridization’s stringency and is specific for each individual gene probe and for each DNA sample.
- In order to remove any unbound probe, a series of washing steps with buffer are then carried out and the membrane is developed, after which the exact location of the probe and its target can be visualised.
- By blotting the DNA and then using a gene probe representing a protein or enzyme from one of the organisms, it is also possible to analyse DNA from different species or organisms. In this way, related genes in different species can be searched for. This technique is generally termed Zoo blotting.
RNA Blotting
- To transfer RNA from gels on to similar membranes, the same basic process of nucleic acid blotting can be used. This enables the identification by hybridisation to a labelled gene probe of specific mRNA sequences of a defined length and is known as Northern blotting.
- It is not only possible to detect specific mRNA molecules with this technique, but it can also be used to quantify the relative amounts of specific mRNA.
- The mRNA transcripts are usually separated under denaturing conditions by gel electrophoresis, since this improves resolution and allows a more accurate estimation of the transcript sizes.
- The format of the blotting may be changed from the transfer of the gel to the direct application to the slots of the nylon membrane-containing specific blotting apparatus.
- This is termed slot or dot blotting and provides a convenient means of measuring the abundance of specific mRNA transcripts without the need for gel electrophoresis; it does not, however, provide information regarding the size of the fragments.
- The ribonuclease protection assay is a further RNA analysis method that overcomes the problems of RNA blotting. Here, the RNA is extracted from a sample and then mixed with a probe representing the solution sequence of interest.
- To form a double-stranded sequence, the probe and the appropriate RNA fragment hybridise. RNAse, which cleaves any single-stranded RNA present but leaves the double-stranded RNA intact, is then added.
- With electrophoresis and an indication of the size of the fragment produced, the intact RNA can then be separated. The effective removal of the RNA background and enhanced sensitivity make the ribonuclease protection assay a popular choice for specific RNA molecules to be analysed.
- The development of RNAi (RNA interference), which inhibits gene expression, was an important step in the RNA analysis field. The degradation of mRNA is promoted here by double-stranded DNA.
- A dicer enzyme cleaves double stranded RNA in the cell, resulting in the formation of small 21-25 bp interfering RNAs (siRNA). The siRNA is complementary to a strand of the target RNA.
- The siRNA guides small RNAi proteins to the appropriate mRNA, where the target is then cleaved and can not be translated. The adoption of this technique in the fields of molecular biology and biotechnology is now beneficial in many areas.
RELATED ARTICLE:
REFERENCES: